Elan Eisenmesser, PhD
Associate Professor

Education
Ph.D., Purdue University, 1998
100 KB
The Eisenmesser lab takes a unique approach to understand protein function, and particularly enzyme function, by utilizing molecular engineering methods to control both structural interactions and the underlying movements that underlie their conformational
changes. The ultimate goal of the Eisenmesser lab is to fully characterize molecular interactions at both atomic resolution and biological levels with a particular emphasis on medically relevant systems that may be exploited to either block
or promote events underlying disease progression. For example, we utilize NMR, X-ray crystallography, and cryo-EM methods together with biochemical and cellular biological studies to fully characterize molecular events that underlie molecular interactions
involved in signaling and cellular homeostasis. Our approaches go well beyond static descriptions of molecular complexes to include the role of dynamics both within enzyme active sites and allosterically coupled movements that modulate function. In
regard to enzyme movements, our studies have been cited as “the first attempt to rationally engineer enzyme motions” (see Peptide Letters, Doucet, 2011) and provide a critical means of unraveling how enzyme movements have evolved to perform
specific tasks.
The current focus of the Eisenmesser lab are three projects described in what follows, which focus on:
- understanding the molecular events of innate immunity that are tied to cancer and inflammatory activities at both the molecular and cellular levels,
- revealing the molecular details of how bacteria and viruses subvert the innate immune system for infection,
- revealing both the structural and dynamic details of critical redox regulators in cellular function and cancer.
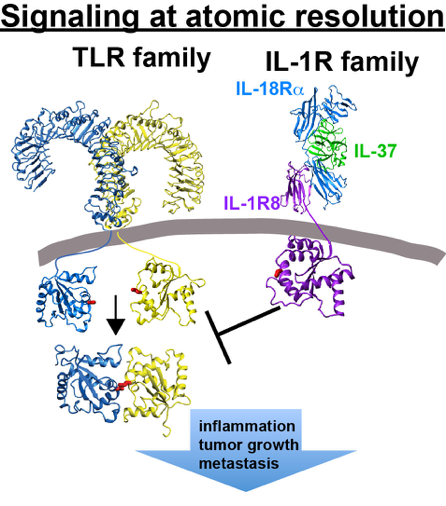
Interleukin signaling on the outside of the cell and receptor interactions on the inside of the cell
Interleukins: Interleukins (ILs) are a major class of cytokines responsible for regulating the innate immune response and their deregulation has been shown to occur during inflammation, cancer, and infection. The Eisenmesser lab is particularly focused on understanding the signaling events utilized by anti-inflammatory interleukins such as IL-37 in order to modulate their structure and biological activities and use as potential therapeutics. Such studies began during the PI’s postdoctoral work on IL-13 but have since been extended to other interleukins that include IL-8 and the anti-inflammatory interleukins that include IL-37 and IL-38.
Toll-Interleukin receptor (TIR) domains: TIR domains are conserved domains found within both interleukin-1 family receptors (IL-1Rs) and Toll-like family receptors, yet despite the 20 years since their discovery, their molecular interactions have eluded direct detection. The Eisenmesser lab is particularly focused on understanding how these TIR domains bridge signaling at the membrane surface with downstream interactions of adaptor TIR domains. While we are interested in determining the molecular interactions that occur at atomic resolution for pro-inflammatory TIRs, such as TLR1/2, we are also interested in understanding how IL-1R family receptors, such as those that include IL-1R8 that work to downregulate inflammation. Finally, we are also interested in understanding how bacteria utilize their own TIR domains to control host cell immunity through host cell TIR mimicry.
- Kendrick AA, Holliday MJ, Isern NG, Zhang F, Camilloni C, Huynh C, Vendruscolo M, Eisenmesser EZ. “The dynamics of interleukin-8 and its interaction with human CXC receptor I peptide.” Protein Sci. (2014) 23(4):464-80.
- Cavalli, G, Justic, JN, Boyle, KE, D’Alessandro, A, Eisenmesser EZ, Herrera, JJ, Hansen, KC, Nemkov, T, Stienstra, R, Garlanda, C, Mantovani, A, Seals, DR, Dagna, L, Joosten, LAB, Ballak, DB, Dinarello, CA. “ Interleukin 37 reverses the metabolic cost of inflammation, increases oxidative respiration and improves exercise tolerance.” PNAS (2017) 114(9):2313-2318.
- Eisenmesser EZ, Gottschlich A, Redzic JS, Paukovich N, Nix JC, Azam T, Zhang L, Zhao R, Kieft JS, The E, Meng X, Dinarello CA. “Interleukin-37 monomer is the active form for reducing innate immunity”. Proc Natl Acad Sci (2019) doi: 10.1073/pnas.1819672116
Covid-19
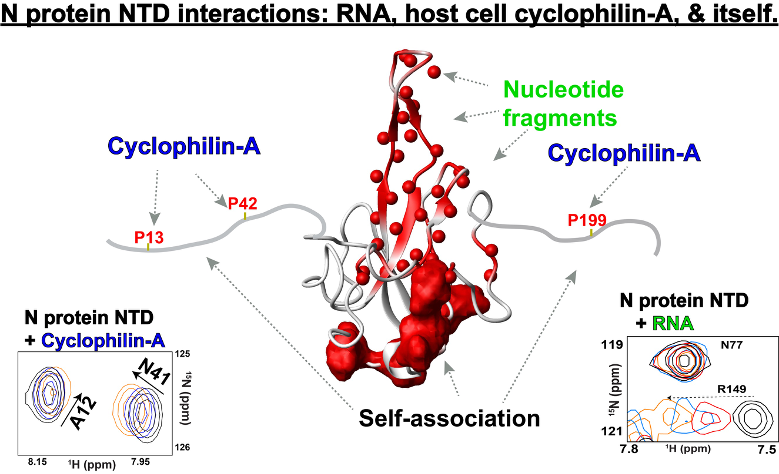
Coronaviruses (CoVs) are evolving pathogens responsible for the recent outbreak of Covid-19 and we have previously studied multiple proteins from viruses that include SARS-CoV-1. While our initial interest was with the nucleocapsid (N) protein from SARS-CoV-1 many years ago, such studies have allowed our group to quickly expand our studies to multiple proteins from SARS-CoV-2 that include its N protein.
The N protein is one of four structural proteins of CoVs and plays a central role in packaging viral RNA and manipulating the host cell machinery, yet its dynamic behavior and promiscuity in nucleotide binding has made standard structural methods to address its atomic-resolution details difficult. To begin addressing the SARS-CoV-2 N protein interactions with both RNA and host cell proteins along with characterizing its unique dynamic behavior, we have specifically focused on the folded N-terminal domain (NTD) of the N protein and its flanking regions using nuclear magnetic resonance solution studies. Our studies reveal a large repertoire of interactions, which includes a temperature-dependent self-association mediated by the disordered flanking regions that also serve as binding sites for host cell cyclophilin-A. In contrast, nucleotide binding is largely mediated by the centrally folded NTD core. The most interesting aspect of this work is that human cyclophilin-A has been known for 10 years to increase viral replication in CoVs through their N protein; however, no known mechanism exists. Our studies pinpoint a potential mechanism of host cell cyclophilin-A in mediating N protein self-association, which is known to modulate cellular condensates responsible for viral replication. Collectively, our studies have begun to reveal the complexities of the nucleocapsid protein dynamics and its preferred interaction sites with its biological targets.
- Clarkson MW, Lei M, Eisenmesser EZ, Labeikovsky W, Redfield A, Kern D “Mesodynamics in the SARS nucleocapsid measured by NMR field cycling” J. Biomol. NMR (2009) 45:217-225.
- Schultz JS, McCarthy MK, Rester C, Sabourin KR, Annen K, DomBourian M, Eisenmesser EZ, Frazer-Abel A, Knight V, Jaenisch T, Morrison TE, Rochford R, Kedl RM. “Development and validation of a multiplex microsphere immunoassay using dried blood spots for SARS-CoV-2 seroprevalence: application in first responders in Colorado, USA.” J. Clin Microbiol. (2021) 61(5):1586-1599. Doi: 10.1128/JCM.00290-21.
- Redzic JS, Lee E, Born A, Issaian A, Henen MA, Nichols P, Blue A, Hansen KC, D'Alessandro A, Vögeli B, Eisenmesser EZ. “The inherent dynamics and interaction sites of the SARS-CoV-2 nucleocapsid N-terminal region”. JMB (2021) Doi:10.1015/j.jmb.2021.167108.
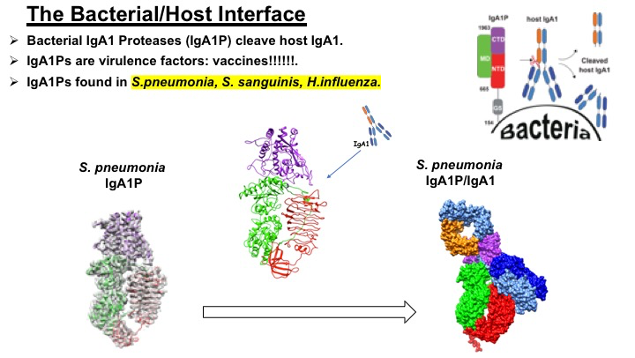
Bacterial/Host Interface
Our interest in human infection and developing strategies to specifically block infection has fostered efforts at understanding how bacterial subvert our initial immune response. CryoEM and NMR studies are focused on determining the molecular mechanisms of giant proteases that have multiple domains, such as the Streptococcus pneumonia IgA1 Protease (IgA1P). These IgA1Ps represent a diverse array of virulence factors that are examples of convergent evolution, as their IgA1P regions are metalloproteases, serine proteases, and cysteine proteases. Many IgA1Ps from different pathogenic bacteria comprise additional domains of unknown function beyond their IgA1P regions. For example, we have solved the structures of small G5 domains from an IgA1P through NMR to begin identifying its ligands. However, our central focus is to elucidate the atomic-resolution details of their IgA1P regions using cryoEM, as illustrated here by our recent elucidations of the ~1300 residue Streptococcus pneumonia IgA1P alone and during catalysis of its IgA1 substrate that indicates a “pakman” like mechanism of active site opening (or “gating”).
- Chi YC, Rahkola JT, Kendrick AA, Holliday MJ, Paukovich N, Roberts TS, Janoff EN, Eisenmesser EZ. “Streptococcus pneumonia IgA1 protease: A metalloprotease that can catalyze in a split manner in vitro.” Protein Sci. (2017) 26(3):600-610.
- Paukovich N, Redzic J, Chi YC, Rahkola JT, Issaian A, Blue A, Hansen KC,
Janoff EN, Eisenmesser EZ. “Streptococcus pneumonia
G5 domains bind different ligands
.” Protein Sci. (2019) 2
8
(10):
1797
-1805.
- Wang Z, Rahkola J, Janoff J, Redzic JS, Chi Y-C, Tran N, Holyoak T, Zheng H, Eisenmesser EZ. “Mechanism and inhibition of Streptococcus pneumoniae IgA1 protease”. Nature Communications (2020) 11(1): 6063-6070 (also featured as Editor’s Highlights https://www.nature.com/collections/hhfigaahch).
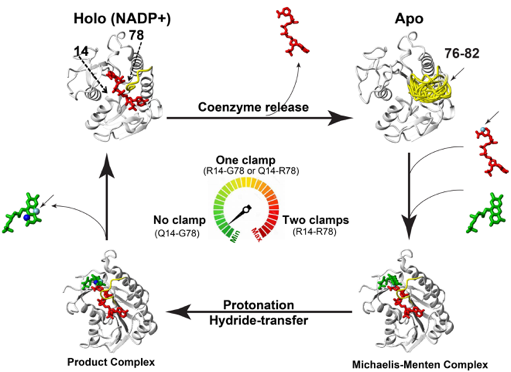
Protein dynamics and function
We have made several major contributions to the field of protein dynamics, which have had direct implications for understanding of protein function, allostery, and have now allowed us to engineer enzyme dynamics in a rational way. The Eisenmesser lab is particularly interested in the interplay between structure and the dynamics that underlie conformational rearrangements of enzymes to direct their function. Our recent studies have focused on a key flavin reductase family of enzymes responsible for modulating cellular redox, called Biliverdin Reductase B (BLVRB), which are so important that they control cellular fate. We have discovered that enzyme dynamics modulate function up to 25 angstroms from the active site in human BLVRB, but we are focused on combining NMR and X-ray crystallography to determine how global motions modulate function in the entire family and how these motions have evolved. Such studies have revealed novel mechanisms of coenzyme regulation, revealing that coenzyme release is the rate-limiting step and that the single hydride modulates the global dynamic and structure.
- Eisenmesser EZ, Capodagli GC, Armstrong GS, Holliday MJ, Isern NG, Zhang F, Pegan SD. “Inherent dynamics within the Crimean-Congo Hemorrhagic fever virus protease are localized to the same region as substrate interactions.” Protein Science (2015) 24(5):651-60.
- Holliday MJ, C, Armstrong GS, Eisenmesser EZ. “Determination of the Fully Catalytic Cycle among Multiple Cyclophilin Family Members and Limitations on the Application of CPMG-RD in Reversible Catalytic Systems”. Biochemistry (2015) 54(38): 5815-27.
- Holliday MJ, Camilloni C, Armstrong GS, Vendruscolo M.,Eisenmesser EZ. “Networks of Dynamic Allostery Regulate Enzyme Function.” Structure (2017) 25(2):276-286.
- Paukovich N, Xue M, Elder JR, Redzic JS, Blue A, Pike H, Miller BG, Pitts TM, Pollock DD, Hansen K, D’alessandro A, Eisenmesser EZ. “Biliverdin Reductase B Dynamics Are Coupled to Coenzyme Binding.” J Mol Biol (2018) 430(18 Pt B):3234-3250.
- Duff M, Redzic JS, Ryan LP, Paukovich N, Zhao R, Nix JC, Pitts TM, Agarwal P, Eisenmesser EZ. “Structure, dynamics, and function of the evolutionarily changing BLVRB family”. J.Biochem. (2020) 168(2):191-202.
- Redzic JR, Duff M, Blue A, Pitts T, Agarwal P, Eisenmesser EZ. “Modulating enzyme function via dynamic allostery within biliverdin reductase B”. Frontiers Molecular Biosciences. 8:691208. Doi:10.3389/fmolb.2021.691208.
Additional studies
The collaborative environment within the Department of Biochemistry & Molecular Genetics has fostered many collaborations, which include those with Dr. Hongin Zheng, Dr. Kirk Hansen, Dr. Angelo D’Alessandro, Dr. Rui Zhao, Dr. Jeff Kieft, and Dr. James DeGregori. For example, multiple ongoing projects with the D’Alessandro lab are focused on key metabolic interactions governed by enzymes and we are focused on producing these enzymes to identify their atomic resolution details that govern their functions.

| Pictures | First Name | Last Name | Job Title | |
|---|---|---|---|---|
| Blaine | Gordon | Postdoctoral Scholar | [email protected] | |
 | Daniel | Rivkin | MIS Student | |
.jpg?sfvrsn=ab43c1b4_0&MaxWidth=120&MaxHeight=160&ScaleUp=false&Quality=High&Method=ResizeFitToAreaArguments&Signature=84FCCD25B06C0AC24A5DDF03B58AF765746AF247) | Gavin | Sparling | Research Service Professional | [email protected] |
 | Jasmina | Redzic | Research Associate | [email protected] |
Interleukins and TIR domains
- Kendrick AA, Holliday MJ, Isern NG, Zhang F, Camilloni C, Huynh C, Vendruscolo M, Eisenmesser EZ. “The dynamics of interleukin-8 and its interaction with human CXC receptor I peptide.” Protein Sci. (2014) 23(4):464-80.
- Cavalli, G, Justic, JN, Boyle, KE, D’Alessandro, A, Eisenmesser EZ, Herrera, JJ, Hansen, KC, Nemkov, T, Stienstra, R, Garlanda, C, Mantovani, A, Seals, DR, Dagna, L, Joosten, LAB, Ballak, DB, Dinarello, CA. “Interleukin 37 reverses the metabolic cost of inflammation, increases oxidative respiration and improves exercise tolerance.” PNAS (2017) 114(9):2313-2318.
- Eisenmesser EZ, Gottschlich A, Redzic JS, Paukovich N, Nix JC, Azam T, Zhang L, Zhao R, Kieft JS, The E, Meng X, Dinarello CA. “Interleukin-37 monomer is the active form for reducing innate immunity”. Proc Natl Acad Sci (2019) doi: 10.1073/pnas.1819672116.
Bacterial infection
- Chi YC, Rahkola JT, Kendrick AA, Holliday MJ, Paukovich N, Roberts TS, Janoff EN, Eisenmesser EZ. “Streptococcus pneumonia IgA1 protease: A metalloprotease that can catalyze in a split manner in vitro.” Protein Sci. (2017) 26(3):600-610.
- Paukovich N, Redzic J, Chi YC, Rahkola JT, Issaian A, Blue A, Hansen KC, Janoff EN, Eisenmesser EZ. “Streptococcus pneumonia G5 domains bind different ligands.” Protein Sci. (2019) 28(10):1797-1805.
Protein dynamics and function
- Eisenmesser EZ, Capodagli GC, Armstrong GS, Holliday MJ, Isern NG, Zhang F, Pegan SD. “Inherent dynamics within the Crimean-Congo Hemorrhagic fever virus protease are localized to the same region as substrate interactions.” Protein Science (2015) 24(5):651-60.
- Holliday MJ, C, Armstrong GS, Eisenmesser EZ. “Determination of the Fully Catalytic Cycle among Multiple Cyclophilin Family Members and Limitations on the Application of CPMG-RD in Reversible Catalytic Systems”. Biochemistry (2015) 54(38): 5815-27.
- Holliday MJ, Camilloni C, Armstrong GS, Vendruscolo M., Eisenmesser EZ. “Networks of Dynamic Allostery Regulate Enzyme Function.” Structure (2017) 25(2):276-286.
- Paukovich N, Xue M, Elder JR, Redzic JS, Blue A, Pike H, Miller BG, Pitts TM, Pollock DD, Hansen K, D’alessandro A, Eisenmesser EZ. “Biliverdin Reductase B Dynamics Are Coupled to Coenzyme Binding.” J Mol Biol (2018) 430(18 Pt B):3234-3250.