Research Projects
Structural Studies of the Eukaryotic Transcription Machinery
Introduction
Before the genetic information stored in the DNA code can be utilized it must be transcribed, in a process in which an RNA copy of the DNA code is made. Our research focuses on structural studies of macromolecular complexes involved in eukaryotic gene expression. Control of a gene expression is mostly effected through the regulation of DNA transcription, which is therefore of utmost significance in cell differentiation and development, and in the response of living cells to their environment. Complex molecular machines comprising many individual proteins perform specific steps of the transcription process. At the center of this complex apparatus is RNA polymerase II (RNAPII), the enzyme responsible for reading the DNA information and synthesizing an RNA copy. In order to carry out transcription and respond to regulatory signals, RNAPII must interact with many other molecules. The objective of our research is to reveal the mechanisms of transcription and its regulation. Electron microscope (EM) images of macromolecular complexes are analyzed to reconstruct their three-dimensional structures, as they appear under conditions similar to those in a living cell. The end result of these efforts will be an understanding of the mechanism of transcription at the molecular level.
RNA polymerase II and the Preinitiation Complex
Transcription initiation requieres the assembly of a large multi-component complex in which RNAPII interacts with a number of accessory proteins known as the general transcription factors (GTFs) to form the preinitiation complex (PIC). The PIC, including nearly 30 different polypeptides, assembles at a promoter and allows RNAPII to recognize the transcription start site. One of the most ambitious goals of our research is to obtain a structure of the entire PIC. A subset of the PIC components including RNAPII, TFIIF, the TATA binding protein (TBP), and TFIIB, constitute the minimum assembly capable of promoter-dependent initiation. We proposed a model for organization of this minimal PIC based on the structure of the RNAPII/TFIIF complex, and work to obtain a structure the minimal PIC is currently underway.
Mediator and Regulation
Mediator, a large complex comprising over 25 different polypetide subunits, is arguably the key player in regulation of transcription in all eukaryotic organisms. Mediator was first characterized in yeast cells, but Mediator homologs have now been identified in all eukaryotes examined. We have generated 3-D structures of yeast, mouse, and human Mediators, as well as a structure of the yeast Mediator/RNAPII (holoenzyme) complex. One of the most intriguing aspects of Mediator structure is that it undergoes a dramatic conformational change upon interaction with RNAPII. This conformational change might play a central role in the regulation mechanism through its effect on the assembly of the PIC.

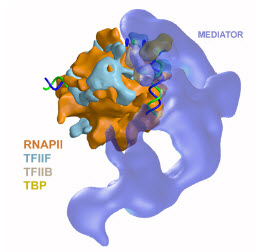
Mediator and the PIC
A variety of activators and repressors interact directly with Mediator which then works as the interface that conveys regulatory information to the basal transcription machinery. Characterization of the Mediator/RNAPII holoenzyme revealed their relative orientation, with Mediator interacting with the back face of polymerase. This is homologous to what has been observed in bacterial cells (where regulatory proteins also interact with polymerase subunits located behind the upstream end of the active site cleft) and suggest the possible conservation of fundamental aspects of the regulation mechanism. While the specific mechanism of regulation by Mediator remains unknown, it seems likely that Mediator might exert an effect on assembly of the PIC.
Future Directions
Structural characterization of the eukaryotic transcription machinery remains the central goal of our research efforts. Cryo-EM studies of yeast and human complexes are currently underway. We are also complementing our EM efforts with biochemical and functional studies preinitiation complex assembly in yeast. We are interested in complementary approaches, such as single-molecule techniques, that could provide additional insight into the mechanism of transcription regulation.