Asturias Lab
Macromolecular Machines
Many essential cellular processes are carried out by "macromolecular machines", large assemblies that often include tens of different proteins, each making a specific contribution to the function of the machine. Characterization of these large macromolecular complexes using a combination of techniques constitutes the next frontier in structural biology.
Interestingly, despite including many different proteins, macromolecular complexes often display minimal enzymatic activity. This suggests that they truly function as macromolecular machines, in which function is related to “mechanical” changes that affect interactions and activity. Characterizing different states and conformations of a macromolecule, and correlating these findings to biochemical and functional information, can be essential to establish the mechanism of these remarkable cellular machines.
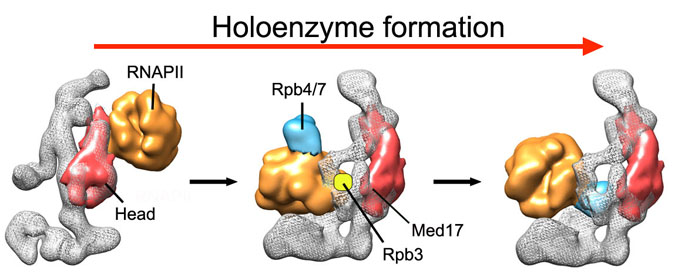
Macromolecular Electron Microscopy
The relative fragility of biological molecules limits the amount of information that can be obtained when some type of radiation is used to interrogate their structure. In X-ray crystallography, signal-to-noise limitations are overcome by obtaining information from very large numbers of identical molecules perfectly arranged in a crystal. However, it is difficult to crystallize a large macromolecular complex and, even if crystals are available, obtaining and interpreting diffraction data is very challenging. In addition, crystallization severely constrains the conformation of a molecule, preventing detection of possible alternate states with functional significance. Macromolecular complexes are also very hard targets for other conventional high-resolution techniques like NMR.
The technique of choice for structural characterization of large (MW > 250,000Da) macromolecular machines is single particle macromolecular electron microscopy (EM). A suitably-recorded electron microscope image of a single large molecule can contain information at near atomic resolution, albeit with very low signal-to-noise ratio. In single particle EM, a very small amount of material is all that is required to record many (103 – 106) images of individual macromolecules quickly preserved from nearly physiological conditions. Information in these images is computationally combined to overcome signal-to-noise limitations and calculate 2D and 3D maps that provide critical information about the structure, molecular organization, and quaternary structure of a macromolecular machine.
Progress in hardware and image processing now make possible routine calculation of accurate macromolecular maps at subnanometer resolution. Interpretation of these maps is frequently aided by considering information from high-resolution structures of individual protein components.