Research
Our group investigates the genetic and biological determinants of diseases that are influenced by the environment. Our previous research focused on the lungs and the immune system, trying to understand why environmental agents cause lung disease and infections in some people but not others. We have discovered that a variant of the gene TLR4 makes some people more susceptible to adverse effects of bacteria, that epigenetic mechanisms may be important in the development of asthma, and that a variant in a mucus producing gene (MUC5B) in the lung markedly enhances the risk of developing pulmonary fibrosis. Understanding the mechanistic basis of how the gain-of-function MUC5B promoter variant leads to development of pulmonary fibrosis and identification of additional variants associated with pulmonary fibrosis are currently a strong focus in the laboratory. We are also studying epigenetic mechanisms that contribute to the development of environmental lung disease. Human cohorts, in vitro studies, and animal models are used to pursue these studies. Our research in these areas has the potential to develop biomarkers for early identification of susceptible individuals, lead to novel concepts about the prevention and pathogenesis of these diseases, and to transform therapy in these diseases.
Genetics and Genomics of Pulmonary Fibrosis
Our pulmonary fibrosis genetics and genomics research seeks to uncover the genetic and environmental determinants of IPF/UIP. Our ultimate goal is to use genetic variants, alone or in combination with gene expression fingerprints and/or peripheral blood
biomarkers, to treat patients based on the genetic subtype of IPF they have, and to initiate treatment in the preclinical stages of disease rather than at a late stage when lung is scarred irreversibly and prognosis is poor. We are focusing on diseases
with the usual interstitial pneumonia (UIP) radiologic and pathologic pattern, specifically, idiopathic pulmonary fibrosis (IPF), chronic hypersensitivity pneumonitis (CHP), and rheumatoid arthritis-associated interstitial lung disease (RA-ILD).
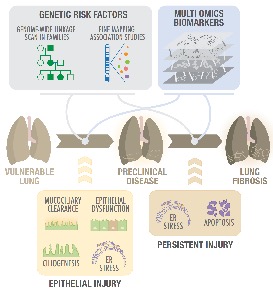
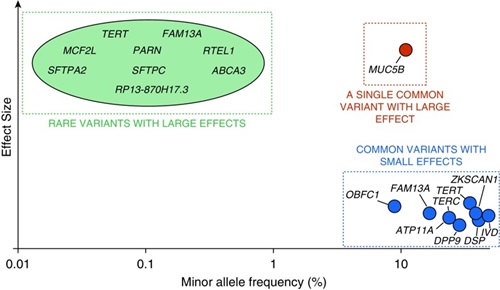
Our work over the past decade has shown that: (1) IIP is a molecularly heterogeneous disease with multiple emerging epigenetic and transcriptional profiles; (2) IIP can be diagnosed preclinically before symptoms and signs of fibrotic lung disease are apparent, and that preclinical pulmonary fibrosis (PrePF) is predictive of radiographic progression and diminished survival; (3) a gain-of-function MUC5B promoter variant is the dominant genetic risk in familial and sporadic IIP, is observed in over 50% of subjects with familial or sporadic IIP, and can be used to identify individuals at risk for PrePF; (4) the gain-of-function MUC5B promoter variant is the dominant genetic risk factor for the development of IPF (OR=5.45; 95%CI=4.91-6.06; P=9.60x10-295), CHP (OR=1.91; 95% CI=1.02-3.59; P=0.045), and RA-ILD (OR=5.5; 95% CI, 4.2=7.3; P=4.7×10-35); and (5) 10 common genetic variants play similar roles in the development of both familial and sporadic IIP.
(Allen and Ortega. Am J Respir Crit Care Med. 2019; 200: 125–127)
Our current work in genetics and genomics of IPF is focused in these areas:
- Comprehensive identification of common and rare genetic variants associated with IPF by whole genome sequencing (WGS) in individuals of European ancestry. We are also exploring how key IPF sequence variants contribute to the etiologic, biologic, and clinical phenotypes of this disease, and examining the generalizability of these genetic risk variants to other ethnic groups.
- Analysis of peripheral blood transcriptome, metabolome, and proteome to identify biomarkers of early disease in subjects with preclinical pulmonary fibrosis. The ultimate goal of this work is to be able preclinical pulmonary fibrosis and to prognosticate among individuals with preclinical pulmonary fibrosis.
- Systems biology integrative analyses of genetic variants, DNA methylation, coding and noncoding transcriptome to identify genetically-anchored genomic signatures of disease. This work will put genetic variants identified in our work in the context of gene regulation and cellular pathways of importance in disease development.
- Use mouse genetics followed by analysis in human cohorts to identify genes and transcripts associated with MUC5B expression that contribute to the development of IPF.
If you are or someone else you know are interested in enrolling in our study, please visit: https://ipfgenetics.org

 Our
rare disease cohort focuses on a critical unmet public health need in interstitial pneumonia, to understand the etiology, natural history, and phenotypic heterogeneity of preclinical pulmonary fibrosis (PrePF), before the lung is scarred irreversibly. Our overall hypothesis is that common genetic variants and environmental risk factors predispose to the development, natural history, and phenotypic heterogeneity of PrePF, and that defining these risk factors will allow us to uncover the common and unique subtypes of PrePF that differ in disease onset and progression. By leveraging our NHLBI-supported discoveries in PrePF, familial interstitial pneumonia (FIP), and idiopathic interstitial pneumonia (IIP), and our NHLBI-supported cohort of FIP families, our project seeks to explain how common genetic variants and
the environment interact to result in the earliest stages of this highly morbid, phenotypically heterogeneous disease. We focus on first-degree relatives (FDRs) previously phenotyped as unaffected (N=2404) from our 1160 FIP families with two or more
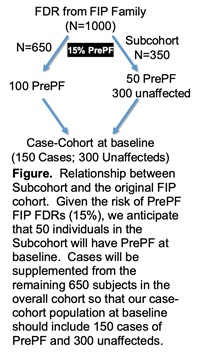
cases of confirmed IIP. Within these 1160 FIP families, we will establish our rare disease cohort of 1000 subjects by selecting up to two asymptomatic, previously phenotyped unaffected FDRs per family. To combine the advantages of our
cohort with the efficiency of a nested case-control study, we will establish a case-cohort study and compare 150 cases of PrePF to 300 unaffected controls (Figure). This approach will allow us to determine the individual and combined contributions
of common genetic variants and environmental features that result in the development of PrePF. By focusing on the natural history of IIP, our results can be used to identify high-risk populations, early forms of the disease, factors associated with
disease progression, and biological targets for drug development. Moreover, a natural history study can also identify critical biomarkers that can be diagnostic of early or established forms of the disease, prognostic of the course of a disease,
predictive of treatment response, or useful in guiding patient selection and dose selection in drug development programs. Our project would establish a prospectively followed high-risk IIP cohort, will identify the genetic and environmental risk factors
for PrePF, and will maximize the utility of this high-risk cohort for ancillary studies focused on primary and secondary prevention of IIP.
Our
rare disease cohort focuses on a critical unmet public health need in interstitial pneumonia, to understand the etiology, natural history, and phenotypic heterogeneity of preclinical pulmonary fibrosis (PrePF), before the lung is scarred irreversibly. Our overall hypothesis is that common genetic variants and environmental risk factors predispose to the development, natural history, and phenotypic heterogeneity of PrePF, and that defining these risk factors will allow us to uncover the common and unique subtypes of PrePF that differ in disease onset and progression. By leveraging our NHLBI-supported discoveries in PrePF, familial interstitial pneumonia (FIP), and idiopathic interstitial pneumonia (IIP), and our NHLBI-supported cohort of FIP families, our project seeks to explain how common genetic variants and
the environment interact to result in the earliest stages of this highly morbid, phenotypically heterogeneous disease. We focus on first-degree relatives (FDRs) previously phenotyped as unaffected (N=2404) from our 1160 FIP families with two or more
cases of confirmed IIP. Within these 1160 FIP families, we will establish our rare disease cohort of 1000 subjects by selecting up to two asymptomatic, previously phenotyped unaffected FDRs per family. To combine the advantages of our
cohort with the efficiency of a nested case-control study, we will establish a case-cohort study and compare 150 cases of PrePF to 300 unaffected controls (Figure). This approach will allow us to determine the individual and combined contributions
of common genetic variants and environmental features that result in the development of PrePF. By focusing on the natural history of IIP, our results can be used to identify high-risk populations, early forms of the disease, factors associated with
disease progression, and biological targets for drug development. Moreover, a natural history study can also identify critical biomarkers that can be diagnostic of early or established forms of the disease, prognostic of the course of a disease,
predictive of treatment response, or useful in guiding patient selection and dose selection in drug development programs. Our project would establish a prospectively followed high-risk IIP cohort, will identify the genetic and environmental risk factors
for PrePF, and will maximize the utility of this high-risk cohort for ancillary studies focused on primary and secondary prevention of IIP.
Regulation of MUC5B in Pulmonary Fibrosis
Gain of function MUC5B variant rs35705950, a G-T transversion, is located ~ 3KB upstream of the transcriptional start site for MUC5B. We have shown that the MUC5B -3 kb upstream region is a classical enhancer, which undergoes an epigenetic transition resulting in accessibility of the underlying DNA for interactions with multiple transcriptional regulators, such as FOXA2. This ultimately leads to RNAP2 loading near the transversion site, enhancer RNA transcription, and MUC5B expression. However, the mechanisms controlling normal and abnormal MUC5B expression and how this relates to aberrant epithelial differentiation in IPF are not fully understood. Our current projects in regulation of MUCB in pulmonary fibrosis are focused in these areas:
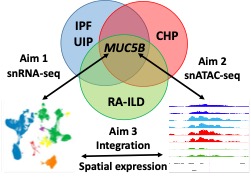
- Single nucleus multi-omic sequencing to understand how the MUC5B promoter variant affects transcriptional regulation and gene expression in clinically distinct types of usual interstitial pneumonia (UIP). By understanding the relationship between the MUC5B promoter variant, transcriptional regulation, and gene expression in IPF, chronic hypersensitivity pneumonitis (CHP), and rheumatoid arthritis associated lung disease (RA-ILD), we will be able to identify the common molecular elements that are critical to the development of UIP. The two critical questions that we plan to address are: 1) what are the common molecular features of UIP, irrespective of clinical context? and 2) does MUC5B promoter variant have a unique molecular signature common to UIP?
Functional Role of MUC5B in Pulmonary Fibrosis
The overall goal of this project is to understand MUC5B regulation and mechanisms linking its functional role in the airway epithelium to fibroproliferation to be able to develop a therapeutic agent that reduces the concentration of MUC5B in the terminal airway of patients with preclinical stages of IPF. We use a combination of animal and cell models to accomplish this goal.
Our key findings to date are: (1) MUC5B mRNA and protein are expressed in bronchiolar epithelia and in IPF honeycomb cysts; (2) MUC5B promoter variant is associated with enhanced MUC5B expression in both unaffected subjects and in patients with IPF; (3) epigenetic mechanisms affect the expression of MUC5B; and (4) in mice, bleomycin induces Muc5b production, and transgenic overexpression of Muc5b results in increased bleomycin-induced lung fibrosis and decreased mucociliary clearance.
There are at least two related hypotheses that link enhanced production of MUC5B in the bronchoalveolar region of the lung to the development of pulmonary fibrosis that we are currently pursuing.
- One line of reasoning focuses on the intracellular relationship between overexpression of MUC5B, metabolic/stress-responsive changes in MUC5B producing cells, the involvement of the respiratory bronchiole, microscopic honeycomb cyst formation, and
repair/regeneration of the distal airspace in IPF. Based on these considerations, as stem cells attempt to regenerate injured bronchiolar and alveolar epithelium, excess expression of MUC5B may disrupt normal developmental pathways and hijack
the normal reparative mechanisms in the distal lung, resulting in chronic fibroproliferation and honeycomb cyst formation.
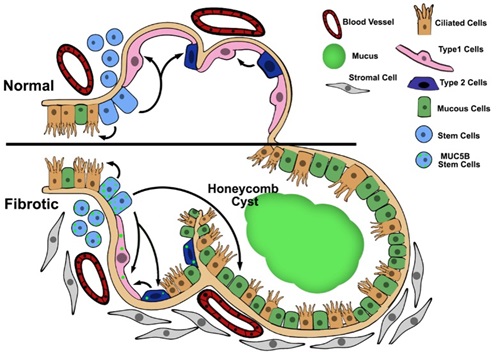
(Evans et al. Physiol Rev. 2016;96:1567-91) - A second line of reasoning focuses on the possibility that IPF is a mucociliary disease caused by recurrent injury/repair at the bronchoalveolar junction, which is initiated and exacerbated by overexpression of MUC5B leading to extracellular effects
of reduced mucociliary clearance, retention of particles, and enhanced lung injury. Based on the relationship between the MUC5B promoter variant rs35705950 and excess production of MUC5B specifically at the bronchoalveolar junction, too
much MUC5B may impair mucociliary function, cause excess retention of inhaled substances (air pollutants, cigarette smoke, microorganisms, etc.), and over time, the foci of lung injury may lead to scar tissue and persistent fibroproliferation
that expands and displaces normal lung tissue.
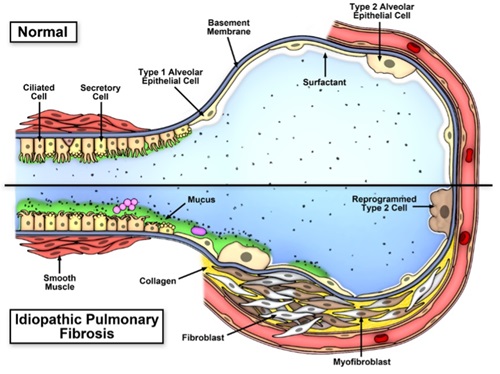
(Evans et al. Physiol Rev. 2016;96:1567-91)
Our current projects in functional role of MUCB in pulmonary fibrosis are focused in these areas:
- Study the effects of MUC5B overexpression on host defense, injury, and repair in cell culture. Airway epithelial cells with and without the MUC5B promoter SNP at air-liquid interface (ALI) are stimulated with pathogen-associated molecular patterns (PAMPs), cigarette smoke condensate (CSC), cytokines, and mechanical stress. Outcomes of interest include MUC5B gene and protein expression, morphologic analysis, mucociliary transport, ER stress/unfolded protein response (UPR), and markers of injury/inflammation.
- Study the effects of MUC5B overexpression on development of fibroproliferative lung disease in mice. Lung injury is induced in Muc5b transgenic and Muc5b deficient mice with bleomycin, cigarette smoke, H1N1 influenza, or other injury models to evaluate differential responses to alveolar injury. Outcomes of interest include mucociliary clearance, cytokine profiles, ER stress/UPR, and lung collagen content (assessed by hydroxyproline assays and second harmonic generation imaging).
- Assessment of various treatment options that reduce Muc5b (mucolytics, antisense oligo sets) in animal models of lung fibrosis.
Role of Ciliogenesis in Pulmonary Fibrosis
The overall goal of this project is to understand the role of increased cilium-associated gene expression, as a consequence of MUC5B overexpression, in the distal airways stem cell (DASC) populations and the contribution of this process to injury/repair and initiation of the fibrotic process in the distal lung. Our transcriptional work demonstrated that, in approximately 40% of patients with IPF, transcripts of cilium genes, MUC5B, MMP7, and the progenitor cell markers KRT14 and KRT5 are highly enriched. The cilium gene signature was validated in multiple lung specimens from the same subjects and in an independent cohort of subjects with IPF, is unique to IPF, and it is associated with the presence of microscopic honeycombing but not fibroblastic foci.
We propose two ways cilia are involved in pathogenesis of IPF. MUC5B overproduction, driven by the MUC5B promoter variant, could lead to impaired cilia-mediated clearance in the small airways; as a result of defective ciliary function, MCC impairment, chronic airway infection and inflammation, bronchiectasis, and distal lung remodeling occur. Furthermore, primary cilia may play role in cellular programming, in particular in distal airway progenitor cells, during repair following injury in the context of MUC5B overproduction
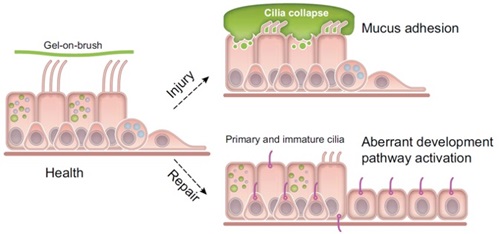
(Evans et al. Physiol Rev. 2016;96:1567-91)
Our current projects in the role of ciliogenesis in pulmonary fibrosis are focused in these areas:
- Use Muc5b transgenic, Muc5b deficient and wild-type animal models to characterize timecourse of multiciliogenesis following injury with bleomycin or H1N1. We are also using animal models with cilia defects (such as conditional deletion of Ift88). Outcomes of interest include expression of multiciliated cell markers, markers of progenitor cells, structural examination of the cilia, mucociliary clearance, and lung collagen content (assessed by hydroxyproline assays and second harmonic generation imaging).
- Study the effect of the MUC5B variant/overexpression on multiciliogenesis in airway-derived organoids from IPF and control lungs. Outcomes of interest include expression of multiciliated cell markers, markers of progenitor cells, structural examination of the cilia, and ciliary beat frequency.
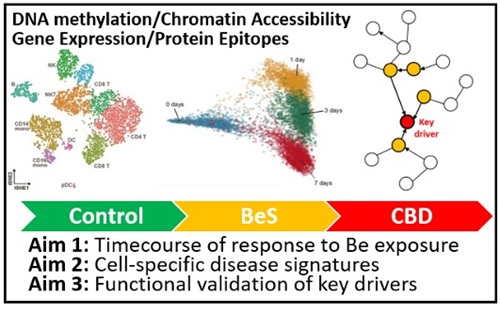
Epigenetics of Chronic Beryllium Disease and Sarcoidosis
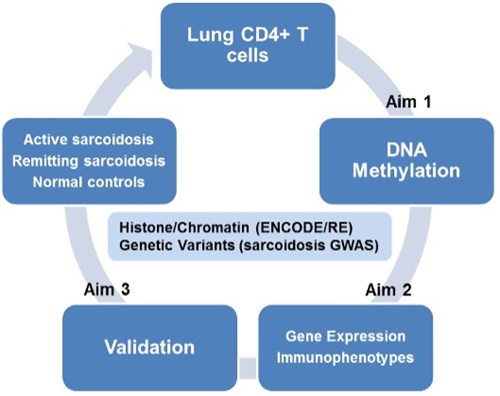
The goal of this project is to define the environmentally induced epigenetic marks and their impact on gene expression that result in the granulomatous lung diseases, chronic beryllium disease (CBD) and sarcoidosis in lung CD4+ T cells. Exposure to an inhaled Be antigen(s) for CBD and unknown antigen(s) for sarcoidosis in the setting of a genetically susceptible host, initiates a Th1 immune response, with antigen presentation occurring via HLA Class II on antigen presenting cell (APC) in the context of CD4+ T cells. Subsequently, CD4+ T cells and APCs are recruited to the lung, proliferate, produce cytokines and chemokines, and eventually form granulomas. In this project, we are defining functional epigenetic loci in by integrative analysis of genomewide DNA methylation and gene expression data, and integration of ENCODE methylation, histone modification, and chromatin accessibility data as well as our GWAS data to prioritize epigenetic marks and networks for confirmation and validation. We are also exploring the potential of methylation marks as biomarkers of disease in peripheral blood and determine if treatment with DNA methylation and demethylating agents affects key immune and regulatory pathways in a second set of CBD and sarcoidosis subjects. We have a recently funded project that aims to define cell-specific regulatory networks and key drivers of cellular response to beryllium (Be) that result in the granulomatous lung disease, chronic beryllium disease (CBD), at the site of organ involvement.
Grant Support
R01HL158668 (Schwartz/Yang)
04/19/2022 – 03/31/2027
NIH-NHLBI
Molecular Determinants of Usual Interstitial Pneumonia (UIP)
R01HL149836 (Schwartz/Clouthier/Yang)
06/01/2020-05/31/2024
NIH-NHLBI
Genes and Transcripts that Interact with MUC5B in Pulmonary Fibrosis
R01ES033678 (Maier/Yang)
02/08/2022-11/30/2026
NIH-NIEHS
Using Multi-Omics to Define Regulators and Drivers of Granulomatous Inflammation and CBD
R01HL140357 (Maier/Yang)
07/01/2018-06/30/2023
NIH-NHLBI
Epigenetic Regulation of Immune Pathways in Sarcoidosis
R01DC019642 (Santos-Cortez/Yang)
09/10/2021-05/31/2026
NIH-NIDCD
Santos-Cortez, Yang (MPI)
Genetic and epigenomic determinants of hearing loss in Hispanic populations
UG3/UH3 HL151865 (Schwartz)
08/01/2020-07/31/2026
NIH-NHLBI
Preclinical Pulmonary Fibrosis, an opportune rare disease cohort
VAMC Merit Review: IO1BX005295 (Schwartz)
10/01/2021-09/30/2025
lncRNAs, Linking genetic susceptibility to molecular phenotype in IPF
W81XWH-16-PRMRP-FPA (PI Schwartz, Project 1 PD Schwartz, Project 4 PD Yang)
Department of Defense
09/30/2017-09/29/2022
Idiopathic Pulmonary Fibrosis, a Disease Initiated by Mucociliary Dysfunction
Milken Institute and Ann Theodore Foundation Breakthrough Sarcoidosis Initiative
Epigenetic Regulation of Severe Pulmonary Sarcoidosis (Li/Maier/Yang)
F32HL154666 NIH/NHLBI (Kim)
01/01/2021 – 06/30/2023
NIH-NHLBI
The role of multiciliated cell dysfunction in pathogenesis of IPF
Lab Address
Schwartz/Yang Laboratory
University of Colorado Denver
12700 East 19th Avenue, 8611
Research Complex 2, Rm 3480
Aurora, CO 80045