Churchill Lab Crystal Structures
Journal Features
 |  |  |
| PMID: 20444609 | PMID: 11024175 |
 |  |  |
| PMID: 12732637 | PMID: 16931575 | PMID: 21125993 |
Structure Gallery
The Asf1 histone H3/H4 complex


EsaI with a modeled acyl-phosphopantetheine
| LasI with modeled C12-acyl chain | |
 |  |
Related Publications
Quorum Sensing (EsaI and LasI): PDB IDs: 1k4j, 1kzf, and 1r05
W.T. Watson, F.V. Murphy IV, T.A. Gould, P. Jambeck, D. L. Val, J. E. Cronan,Jr., S. Beck von Bodman, and M.E.A. Churchill* ³Crystallization and Rhenium
MAD Phasing of the Acyl-homoserinelactone Synthase EsaI.² (2001) Acta Crystallographica D57, 1945-1949.
W.T. Watson, T. D. Minogue, Dale L. Val, S. Beck von Bodman, and M.E.A. Churchill* (2002) ³Structural Basis and Specificity
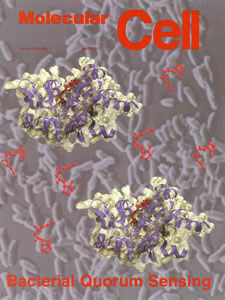
of Acyl-homoserine lactone Signal Production in Bacterial Quorum Sensing² (2002) Molecular Cell 9, 685-694 (with cover photo).
T.A. Gould, W.T. Watson, K.-H. Choi, H.P. Schweizer, and M.E.A. Churchill* ³Crystallization of
Pseudomonas aeruginosa AHL synthase LasI using beta-turn crystal engineering.² (2004) Acta Cryst. D60, 518-520.
T.A. Gould, H.P. Schweizer, and M.E.A. Churchill* ³Structure of the Pseudomonas aeruginosa Acyl-homoserinelactone
Synthase LasI.² (2004) Molecular Microbiology 53, 1135-1146.
| HMGD + DNA | DNA Intercalation by HMGD |
HMGD bends DNA |
|

| pairs of HMG domains bind DNA | The tail of HMGD binds in the DNA major groove |
|  |
Related Publications
Pairs of HMG domains: PDB IDs: 1 hma, 1qrv and 3nm9
D.N.M. Jones, M.A. Searles, G.L. Shaw, M.E.A. Churchill, S.S. Ner, J. Keeler, A.A. Travers, & D. Neuhaus* ³The Solution Structure and Dynamics of the DNA-binding Domain of HMG-D from Drosophila Melanogaster.² (1994) Structure 2, 609-627.
M.E.A. Churchill*, D.N.M. Jones, T. Glaser, H. Hefner, M.A. Searles, & A.A. Travers ³HMG-D is an Architecture-Specific Protein that Preferentially Binds to DNA Containing the Dinucleotide TG.² (1995) EMBO J. 14, 1264-1275 [PMC 398206].
F.V. Murphy IV, J. V. Sehy, Y.G. Gao, L.K. Dow & M.E.A. Churchill* ³Co-crystallization and Preliminary Crystallographic Analysis of the HMG Domain of HMG-D Bound to DNA.² (1999) Acta Crystallographica D55, 1594-1597.
F.V. Murphy IV, R. M. Sweet, & M.E.A. Churchill* ³The Structure of a Chromosomal High-Mobility-Group Protein-DNA Complex Reveals Sequence-neutral Mechanisms Important for Non-sequence-specific DNA Recognition.² (1999) EMBO J. 18, 6610-6618 [PMC 1171724].
L.K. Dow, D.N.M. Jones, S.A. Wolfe, G.L. Verdine, and M.E.A. Churchill* ³Structural Studies of the High Mobility Group Globular Domain and Basic Tail of HMG-D Bound to Disulfide Cross-linked DNA² (2000) Biochemistry 39, 9725-9736.
J. Klass, F.V. Murphy IV, S. Fouts, M. Serenil, A. Changela, J. Siple, and M.E.A. Churchill* ³The Role of Intercalating Residues in Chromosomal High-Mobility-Group Protein DNA Binding, Bending and Specificity.² (2003) Nucleic Acids Research 31, 2852-64 [PMC 156723].
M.E.A. Churchill*, J. Klass, and D.L. Zoetewey ³Structural Analysis of HMGD-DNA Complexes Reveal Influence of Intercalation on Sequence Selectivity and DNA Bending ² (2010) J. Molecular Biology
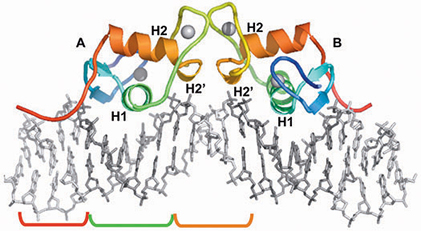
Progesterone Receptor Bound to DNA
Related Publications
Progesterone Receptor-DNA complex: PDB ID 2c7a
S.C. Roemer, D. Donham, L. Sherman, V.H. Pon, D.P. Edwards, and M.E.A. Churchill* ³Structure of the Progesterone Receptor-DNA complex: Novel Interactions Required for Binding to Half-site Response Elements² (2006) Molecular Endocrinology 20, 3042-3052 (with cover photo) [PMC 2532839].
S.C. Roemer, J. Adelman, M.E.A. Churchill*, and D.P. Edwards* ³Mechanism of high mobility group protein B enhancement of progesterone receptor sequence specific DNA binding.² (2008) Nucleic Acids Research, 36, 3655-3666 [PMC 2441811].
K.K. Hill, S.C. Roemer, D.N.M. Jones, M.E.A. Churchill* and D.P. Edwards* ³The progesterone receptor coactivator JDP2 mediates activity through interaction with residues in the carboxyl terminal extension of the DNA binding domain² (2009) J. Biological Chemistry 284, 24415-24424 (paper of the week) [PMC ].
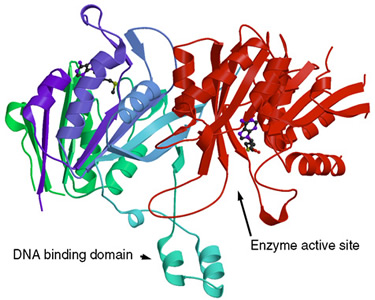
RsrI DNA methyltransferase
Related Publications
RsrI: PDB IDs: 1eg2, 1nw5, 1nw6, 1nw7, and 1nw8
R.D. Scavetta, C.B. Thomas, M.A. Walsh, S. Szegedi, A. Joachimiak, R.I. Gumport, M.E.A. Churchill* ³Structure of RsrI methyltransferase, a member of the N6-Adenine beta class of DNA methyltransferases² (2000) Nucleic Acids Research 28, 3950-3961 (with cover photo) [PMC 110776].
C.B. Thomas, R.D. Scavetta, R.I. Gumport, and M.E.A. Churchill* ³Structures of liganded and unliganded RsrI N6-Adenine DNA methyltransferase: a distinct orientation for active cofactor binding² (2003) J. Biological Chemistry 278, 26094-26101 (with cover photo).


