Alessandra Brambati, PhD
Assistant Professor
Contact Information:
University of Colorado Denver
Department of Pharmacology
Mail Stop 8303, RC1-North
12800 East 19th Ave
Aurora CO 80045
Phone:
Fax: (303) 724-6124
Email: [email protected]
Office: RC1-South, L18-6113
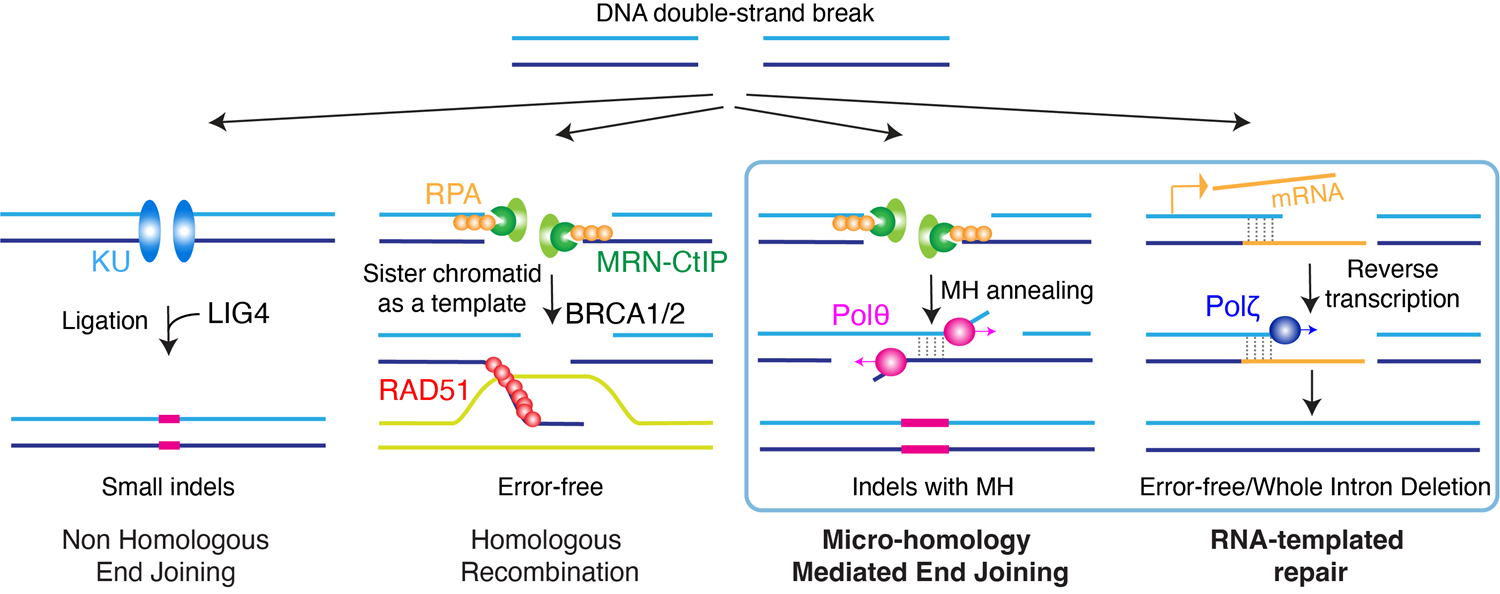
We study the molecular mechanisms that regulate DNA repair and genome instability, focusing on non-canonical DNA double-strand break (DSB) repair pathways. DSBs are one of the most toxic forms of DNA damage. If left unrepaired or repaired inaccurately, DSBs can lead to mutations, chromosomal rearrangements, and even cell death. Effective DSB repair is therefore essential for preserving genomic stability and preventing a range of diseases and cellular malfunctions. The canonical pathways for repairing DSBs are non-homologous end joining (NHEJ) and homologous recombination (HR) pathways. NHEJ repairs breaks by sealing DNA ends, often causing small insertions or deletions, while HR uses the sister chromatid as a template, making it error-free. In the lab we study two non-canonical and poorly characterized DSB repair pathways: microhomology-mediated end-joining (MMEJ) and RNA-templated DSB repair (RT-DSBR). Our research explores how disruptions in these critical processes contribute to disease development and drive evolutionary change.