Benjamin Scholl
Assistant Professor
/benjamin-schollc7798be8302864d9a5bfff0a001ce385.jpeg?sfvrsn=851efb4_0)
Department of Physiology and Biophysics
University of Colorado Anschutz
RC1 North Tower P18-7104
Mail Stop 8307
Aurora, CO 80045
Tel (971) 645-5409
E-mail: [email protected]
External Website: https://benjaminscholl.com/
Graduate Program Affiliations:
Neuroscience Graduate ProgramBioengineering Graduate Program
Medical Science Training Program; Biomedical Science Program
Co-Appointment in Department of Opthamology
Scholl_cv_current
(pdf)
89 KB
Shared Content Block:
Physiology & Biophysics Styles -- Borderless table for directories
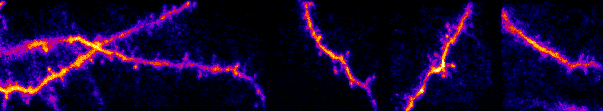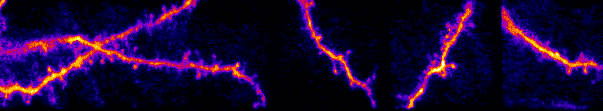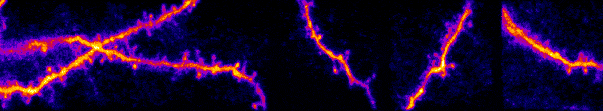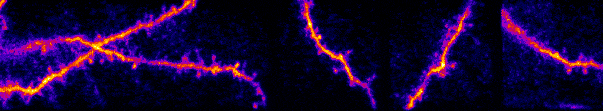
My lab studies synaptic networks. Each neuron receives a wide collection of inputs. These inputs can drive, suppress, or subtly modulate their target. The suite of operations neurons can perform are defined by these inputs and their dynamics. Outside network dynamics, the only way a neuron can dramatically change its operational capacity is through plasticity (e.g. learning, development) of these inputs. Thus, to understand how individual neurons and circuits transform information, we must build a fundamental understanding of synaptic networks. I believe this knowledge will extend beyond fundamentals and lead to novel insights into circuits in neurological and developmental disorders.
Our work uses a variety of techniques to study sensory processing within single cells and across large-scale populations in vivo. Historically this has been with electrophysiology (intracellular and extracellular recordings) and multiphoton calcium imaging. We have expanded to two-photon optogenetics, functional connectomics with electron microscopy, novel viral constructs, and gene editing to disrupt naturally expressed proteins and receptors.
For current and ongoing projects please see lab webpage: www.benjaminscholl.com
/240417penn_1113-(1).jpg?sfvrsn=99ced3bb_1/
240417penn_1113-(1)
.jpg)
| Joe Barreto Professional Research Assistant |
/181611375_emilydale-2025-headshot.jpg?sfvrsn=574917b4_0) | Emily Dale Research Assistant |
Alfred P. Sloan Research Fellow (2023)
Brain Research Foundation Seed Grant Award (2023)
IDDRC Program Development Award (2023)
University Research Foundation Award (2023)
Whitehall Foundation Award (2022)
VRC Pilot Award (2021)
McCabe Fund Award (2021)
Allison Doupe Fellowship Award (2019)
MPFI Scientific Achievement (2018)
Neurizons Young Investigator (2016)
NSF Graduate Research Fellowship Honors (2012)
United States Representative to Nobel Laureate meeting for Physiology and Medicine (2011)
Thomas C, Ryan M, McNabb MC, Kamasawa N, and Scholl B (2024) Astrocyte coverage of excitatory synapse correlates to measures of synaptic structure and function in primary visual cortex. GLIA. https://onlinelibrary.wiley.com/doi/full/10.1002/glia.24582
Thomas C, Ryan M, Kamasawa N, and Scholl B (2023) Postsynaptic mitochondria are positioned to support functional diversity of dendritic spines. eLife. https://elifesciences.org/reviewed-preprints/89682
Yates JL and Scholl B (2022) Unraveling functional diversity of cortical synaptic architecture through the lens of population coding. Frontiers in Synaptic Neuroscience. https://doi.org/10.3389/fnsyn.2022.888214
Scholl B*, Tepohl C*, Thomas C, Ryan M, Kamasawa N and Fitzpatrick D (2022) A Binocular synaptic network supports interocular response alignment in visual cortical neurons. Neuron. https://doi.org/10.1016/j.neuron.2022.01.023
Scholl B*, Thomas C*, Ryan M, Kamasawa N and Fitzpatrick D (2021) Cortical neuron response selectivity derives from strength in numbers of synapses. Nature. https://www.nature.com/articles/s41586-020-03044-3
Wilson DE*, Scholl B* and Fitzpatrick D (2018) Differential tuning of excitation and inhibition underlies direction selectivity in ferret visual cortex. Nature.https://www.nature.com/articles/s41586-018-0354-1