
Jing Hong Wang, MD, PhD
Associate Professor of Immunology & Microbiology, University of Colorado Anschutz School of Medicine
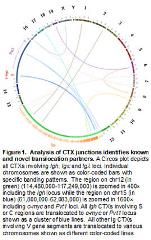 Immune Evasion Mechanism and Cancer Immunotherapy in B cell lymphomas. The goal of our laboratory is to understand the molecular mechanisms that promote chromosomal translocations and cancer development in B lymphocytes by employing mouse models. In particular, we focus on DNA double strand breaks (DSBs) response factors. These proteins are essential for properly repairing DSBs. In the absence of these factors, DSBs will separate and progress into chromosomal breaks and translocations (Chen Z et al., 2016) (Figure 1). We are investigating how these proteins suppress oncogenic translocations or cancer development with cutting-edge techniques such as next generation sequencing and single cell RNA-seq.
Immune Evasion Mechanism and Cancer Immunotherapy in B cell lymphomas. The goal of our laboratory is to understand the molecular mechanisms that promote chromosomal translocations and cancer development in B lymphocytes by employing mouse models. In particular, we focus on DNA double strand breaks (DSBs) response factors. These proteins are essential for properly repairing DSBs. In the absence of these factors, DSBs will separate and progress into chromosomal breaks and translocations (Chen Z et al., 2016) (Figure 1). We are investigating how these proteins suppress oncogenic translocations or cancer development with cutting-edge techniques such as next generation sequencing and single cell RNA-seq.
In addition, we investigate how the interplay of DNA damage and immune response regulates the mechanisms of cancer immune evasion. We have established interesting B cell lymphoma models which harbor defects in DNA repair and appear to evade the recognition of immune cells. We will employ these unique models to elucidate how defects in DNA damage responses enable cancer cells to evade the recognition of immune systems. Furthermore, we are interested in developing novel cancer immunotherapy to treat B cell lymphomas.
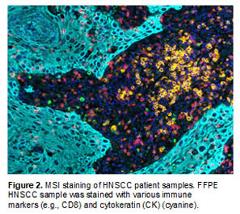 Mechanisms of Immune Evasion in Head Neck Squamous Carcinomas (HNSCC). We have established transplanted HNSCC models which evade the recognition of CD8 T cells and NK cells (Mishra AK, et al., 2016). In addition, we are building other spontaneous or transplanted HNSCC models in which common tumor suppressors or oncogenes of HNSCCs are disrupted. We will employ these unique mouse models and human HNSCC samples to elucidate how HNSCC cancer cells evade the recognition of immune systems. To do so, we are using cutting-edge techniques such as multiplex spectra imaging (MSI) (Figure 2) and mass cytometry (CyTOF). In addition, we are interested in developing novel cancer immunotherapy to more effectively treat HNSCCs.
Mechanisms of Immune Evasion in Head Neck Squamous Carcinomas (HNSCC). We have established transplanted HNSCC models which evade the recognition of CD8 T cells and NK cells (Mishra AK, et al., 2016). In addition, we are building other spontaneous or transplanted HNSCC models in which common tumor suppressors or oncogenes of HNSCCs are disrupted. We will employ these unique mouse models and human HNSCC samples to elucidate how HNSCC cancer cells evade the recognition of immune systems. To do so, we are using cutting-edge techniques such as multiplex spectra imaging (MSI) (Figure 2) and mass cytometry (CyTOF). In addition, we are interested in developing novel cancer immunotherapy to more effectively treat HNSCCs.
Molecular mechanism of class switch recombination: More than 90% of human lymphomas are B cell-derived. This is likely because B cells extensively shuffle their genomic DNA during immune responses (Chen Z and Wang JH, 2014). Upon immunizations or infections, activated B cells undergo somatic hypermutation (SHM) and class switch recombination (CSR). Both SHM and CSR rely on the activity of an enzyme called activation induced deaminase (AID). In activated B lymphocytes, AID causes point mutations in the variable regions of the immunoglobulin (Ig) genes. These mutations cause small changes in the protein sequence of antibodies and lead to increased antibody affinity for specific antigens. AID also induces DSBs in the repetitive switch regions that lie upstream of each constant region exon of the Ig heavy chain locus to mediate CSR. CSR assigns antigen-appropriate effector functions to antibody molecules and is, therefore, a process important for productive immune responses against pathogens. One of our research interests is to elucidate the mechanisms regulating the signaling control of CSR process. B cells express phosphatase and tensin homolog (PTEN) and multiple isoforms of class IA phosphoinositide 3-kinase (PI3K) catalytic subunits, including p110α and p110δ, whose roles in CSR remain unknown or controversial. Our study (Chen Z et al., 2015) reveals that a signaling balance between PTEN and PI3K isoforms is essential to maintain normal CSR. Furthermore, we discovered an important role of p110α in suppressing CSR and found that AKT is the essential downstream effector kinase of PI3Ks to mediate CSR inhibition (Chen Z et al., 2015). We also propose to address how antigen receptor and co-receptors on B cells cooperatively regulate CSR via affecting AID expression.
Education
2002.9-2004.6 - Ph.D. Pathology Program, University of Chicago, Chicago, IL. Degree was granted in June, 2004. Thesis Advisor: Dr. Yang-Xin Fu
1996.9-1998.12 - Master of Science. Graduate Program in Department of Molecular Genetics, Ohio State University, Columbus, OH. Degree was granted in December, 1998. Advisor: Dr. Berl Oakley
1991.8-1996.7 - Bachelor of Medicine (equivalent to M.D. in U.S.). Department of Basic Medical Sciences, Beijing Medical University, Beijing, P.R. China. Degree was granted in July, 1996.
Positions
2016.7 - present - Associate Professor, Department of Immunology and Microbiology, University of Colorado School of Medicine
2011.1-2016.6 - Assistant Professor, Department of Immunology and Microbiology, University of Colorado School of Medicine
2009.6-2010.12 - Instructor of Pediatrics, Immune Disease Institute, Children’s Hospital Boston, Harvard Medical School. Lab PI: Dr. Frederick W. Alt.
2004.7-2009.6 - Postdoctoral Research Fellow, Immune Disease Institute, Children’s Hospital Boston, Harvard Medical School. Lab PI: Dr. Frederick W. Alt.
1999.1-2002.8 - Research Associate, Department of Pathology, University of Chicago. Lab PI: Dr. Yang-Xin Fu.
Honors/Awards
2017: AAI Lustgarten-Thermo Fisher Scientific Memorial Award
2014: AAI 2014Early Career Faculty Travel Award
2012: Outstanding Junior Faculty Award, National Jewish Health
2012-2015: American Society of Hematology (ASH) Scholar Award
2011-2014: Boettcher Foundation Webb-Waring Early Career Investigator Award
2005-2008: Leukemia and Lymphoma Society (LLS) senior fellowship
1996-1997: Distinguished University Fellowship, Ohio State University
1996: GuangHua Scholarship, Beijing Medical University
| Zhangguo Chen, PhD Assistant Research Professor zhangguo.chen@cuanschutz.edu | |
| Rachel A. Woolaver Graduate Student, Immunology Program rachel.woolaver@cuanschutz.edu | |
Alexandra Krinsky Professional Research Assistant (PRA) | |
Samantha Chen Postdoctoral Fellow | |
Jessy John Postdoctoral Fellow |
Immunology Microbiology (SOM)
CU Anschutz
Research I North
12800 East 19th Avenue
Mail Stop 8333
Aurora, CO 80045
303-724-4224