
R. Lee Reinhardt, PhD
Associate Professor, Department of Immunology and Genomic Medicine, National Jewish Health
Associate Professor, Department of Immunology & Microbiology, University of Colorado SOM
 [email protected]
[email protected]
Adaptive Immunity, Innate Immunity, mucosal Immunology, Allergic disease, Pulmonary disease, Host-Pathogen Interactions (parasites), Human/Mouse models, Space Biology
Our research program focuses on understanding how T cells and innate lymphoid cells in mucosal tissues orchestrate type-2 inflammation to protect against infection and suppress allergic and autoimmune disease. We are also interested in how microgravity and exposure to space/lunar environments impact pulmonary health and immunity.
The Reinhardt Lab is committed to mentoring and promoting DEI. Dr. Reinhardt was selected as the recipient of the Duke School of Medicine Gordon Hammes Faculty Award "In recognition of his abiding commitment to excellence in graduate teaching and for hisexemplary leadership in the mentoring of graduate and undergraduate students". He continues to hone his mentoring style to best engage each individual student (ex. NIH's 5-part mentoring course: Becoming a Resilient Scientist; Bystander/Upstander training).
The long-term goal of our research is to expand our understanding of the following five areas: 1) The cellular and molecular events that influence allergic disease susceptibility and initiation; 2) The mechanisms regulating immunity to parasites as a foundation for vaccine development; 3) The role and relationship between follicular T helper (Tfh), T-helper 2 (Th2), and follicular regulatory (Tfr) cells in the development/suppression of allergic and infectious disease, 4) The role of group 2 innate lymphoid cell (ILC2) subsets in mucosal barrier immunity, and 5) The mechanisms driving interferon-mediated autoinflammatory diseases.
Our work has largely focused on understanding the generation, function, and cell fate decisions of CD4+ T-helper (Th) cell subsets in vivo following infection. To do this, we make use of various cytokine and transcription factor reporter mouse models to elucidate Th1, Th2, Tfh, and Tfr development and function. These models provide the opportunity to visualize immune cell function directly in vivo using flow cytometry in combination with conventional immunofluorescence (Figure 1) and live tissue imaging using multi-photon microscopy (Figure 2).
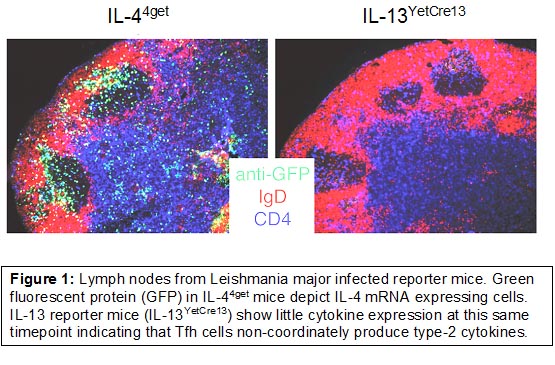
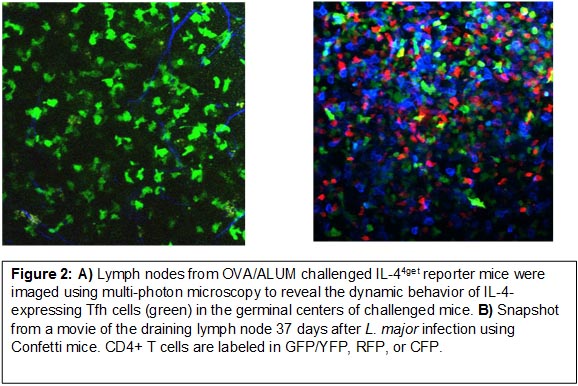
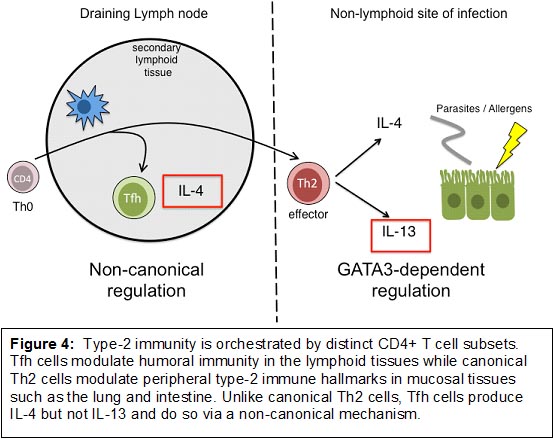
Together, these studies have provided the basis to understand an unexpected, yet important bifurcation in type-2 immunity orchestrated by follicular T cells in the lymphoid tissues and Th2 cells in mucosal non-lymphoid tissues (Figure 4).
Beyond visualizing cellular function, these reporter animals provide excellent tools to study gene expression and function as well as epigenetic gene regulation in physiologic settings. Specifically, these reporter systems allow us to isolate rare populations of innate and adaptive immune cells from various tissues to explore their transcriptional programs and epigenetic profiles at various times during an immune response. We have now established collaborations to continue to investigate these cellular programs and epigenetic events at the single cell level.
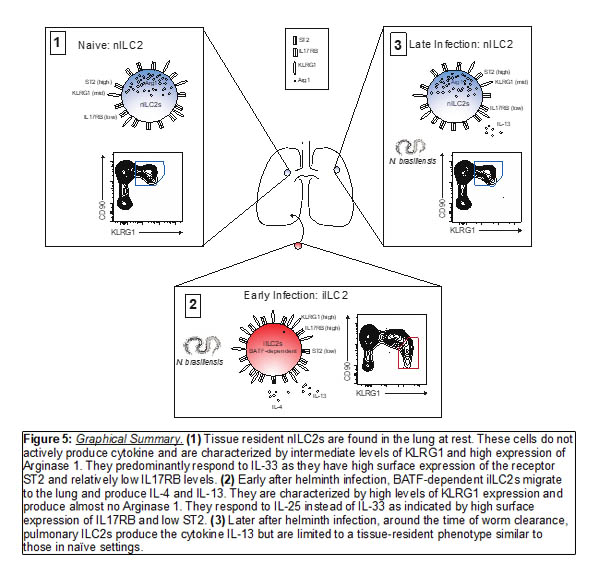
We have recently expanded our studies of Type-2 immunity into innate lymphoid cells, specifically ILC2s. Our most recent publication described the transcription factor, BATF, as being required for the migratory iILC2 subset which traffics to the lung early after helminth infection and produces type 2 cytokines important in tissue repair (http://immunology.sciencemag.org/cgi/content/full/5/43/eaay3994?ijkey=WjdGW8JiShChg&keytype=ref&siteid=immunology) (Figure 5). We are interested in further understanding where this migratory iILC2 subset originates, the mechanism of trafficking, and how it contributes to the long-term immune landscape of the lung. In addition to murine ILC2 studies, we are also investigating these cells in human lung tissue.
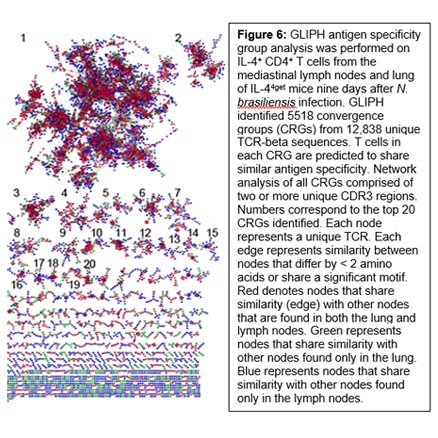
 We are also very interested in anti-helminth immunity. Currently, the breadth, depth, and antigen-reactivity of the responding CD4+ T cell repertoire after helminth infection is largely unknown, and few studies to date have investigated in depth the responding T cell repertoire. How the anti-helminth T cell repertoire changes upon secondary infection, whether it differs among lymphoid and nonlymphoid tissues, and the uniqueness of the repertoire between Tfh and Th2 cell subsets remains unexplored. Our data suggest that the anti-helminth T cell repertoire is diverse and with broad antigen-specificity. This diversity makes the response to these large extracellular worms unique from what is observed in responses to dominant epitopes found in viral or bacterial immunity. It also complicates our understanding of vaccine epitope discovery. We hope to leverage a better understanding of the natural immune repertoire into the development of more effective and safe vaccine.
We are also very interested in anti-helminth immunity. Currently, the breadth, depth, and antigen-reactivity of the responding CD4+ T cell repertoire after helminth infection is largely unknown, and few studies to date have investigated in depth the responding T cell repertoire. How the anti-helminth T cell repertoire changes upon secondary infection, whether it differs among lymphoid and nonlymphoid tissues, and the uniqueness of the repertoire between Tfh and Th2 cell subsets remains unexplored. Our data suggest that the anti-helminth T cell repertoire is diverse and with broad antigen-specificity. This diversity makes the response to these large extracellular worms unique from what is observed in responses to dominant epitopes found in viral or bacterial immunity. It also complicates our understanding of vaccine epitope discovery. We hope to leverage a better understanding of the natural immune repertoire into the development of more effective and safe vaccine.
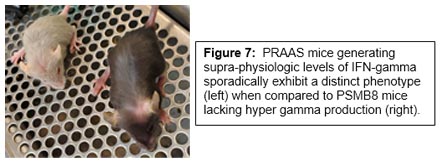
Another area we are excited about involves our efforts to understand IFN-gamma driven autoinflammatory diseases with the goal of modeling rare childhood syndromes. We have generated a new mouse model harboring a human mutation present in rare proteasome-associated autoinflammatory syndromes (PRAAS) (Figure 6). This is the first PRAAS-specific mouse model, and this pre-clinical model will allow us to better understand how type-1 and type-2 interferons are involved in disease pathogenesis. We believe mutations in subunits of the immunoproteasome create a pathogenic feedback loop where chronic production of IFNs promote increased inflammation and pathology.
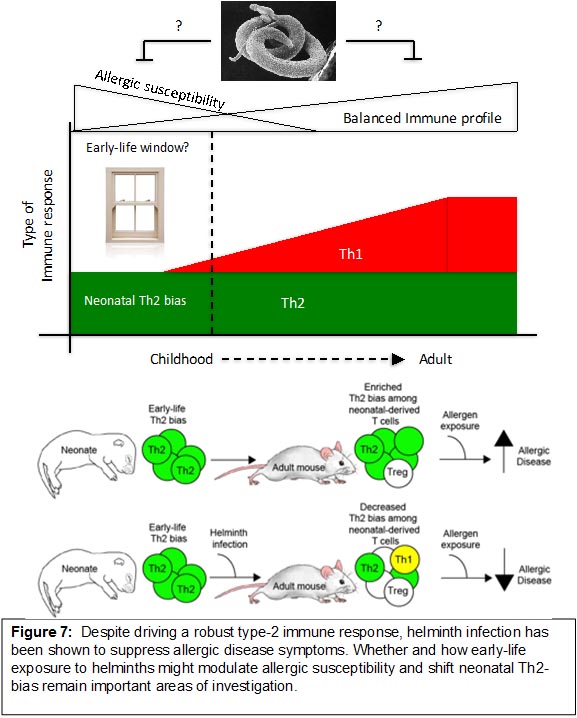
Lastly, we are exploring various mechanisms related to how early-life helminth exposure can prevent or ameliorate allergic disease symptoms (Figure 8). The goal is to uncover novel mechanisms that can be leveraged into therapies that mimic the beneficial effects of helminths on allergic disease susceptibility.
Current Lab Members
| Postdocs and Fellows: | |
 | Mindy M Miller, PhD I received my PhD from the University of Missouri where I investigated neonatal CD4 T cell responses under the guidance of Dr. Habib Zaghouani. This work introduced me to the concept that immune responses in early life can profoundly shape the development of diseases later on. Along these lines, in the Reinhardt Laboratory I am currently investigating how helminth infection imprints a protective function on macrophages to prevent or alleviate allergic asthma. In addition, my research has focused on the functional characterization of a novel subset of migratory ILC2 cells that are critical in early barrier immunity. |
| Research Scientists: | |
 | Ivy K Brown, MSc (Sr. Researcher/Lab Manager) I obtained my master’s degree in Molecular Biology specialized in human health at the Free University of Brussels in Belgium. My current research projects primarily focus on characterizing various mouse models of relevant human diseases, as well as various other projects in which the lab is invested. We aim to investigate the cellular mechanisms behind these disorders with an ultimate goal of developing therapies for treatment. I am also responsible for lab management and all the duties encompassed therein. |
Past Lab Members: (current position)
Postdocs and Fellows:
Thomas O’Brien (Beckman-Coulter)
Preeyam Patel (Postdoc New York University)
Graduate Students:
Katherine Bao (Postdoc Genentech)
Mark Dell’Aringa (Postdoc National Jewish Health)
Research Scientists/ Lab Managers:
Ann Miller (Duke University)
Julio Barrera (Duke University)
Undergraduate Students:
Jingxiao Jin (Baylor Medical School)
Simon Jiang (Duke University; Element Genomics)
Early Career Scientists/ Pre-collegiate Students:
Rebecca Shen (Stanford)
Reinhardt Lab (Winter 2020)
Lee, Otto and Mindy – Science Immunology publication commemoration!

Reinhardt Lab (Winter 2019)
Lee, Ivy and Mindy – Happy holidays!

Reinhardt Lab (Summer 2019)
Mindy, Mary, Ivy and Lee - Winners of the annual retreat costume contest!

Reinhardt Lab (Summer 2018)
Mindy, Ivy, and Mary on Independence Day

Reinhardt Lab (Winter 2017)
Mark, Preeyam, and Ivy - Welcome to Denver!

Reinhardt Lab (Winter 2014)
Tommy, Mark, Katherine, Jingxiao and Ann (not pictured) –
The Fantastic First Five

Journal Articles: (* denotes equal contribution)
- Miller MM, Patel PS, Bao K, Danhorn T, O'Connor BP, Reinhardt RL. (2020) BATF acts as an essential regulator of IL-25-responsive migratory ILC2 cell fate and function. Science Immunology. Jan 10;5(43).
- Dell’Aringa M and Reinhardt RL (2018) Notch signaling represents an important checkpoint between follicular T-helper and canonical T-helper 2 cell fate. Mucosal Immunology 11(4):1079-1091
- Bao K, Carr T, Wu J, Barclay W, Jin J, Ciofani M, Reinhardt RL (2016) BATF modulates the Th2 locus control region and regulates CD4+ T cell fate during anti-helminth immunity. J Immunol 197 (11):4371-4381
- O’Brien TF, Bao K, Ang W.X.G, Abraham S, Reinhardt RL (2016) L-4 and IL-13 expression by Natural Killer T cells is tightly regulated throughout development and in settings of allergic inflammation. Mucosal Immunology 9(3):597-609
- Reinhardt RL, Liang HE, Bao K, Price AE, Mohrs M, Kelly BL, Locksley RM (2015) A novel model for IFN-gamma-mediated autoinflammatory syndromes. J Immunol 194(5):2358-68
- Liang HE*, Reinhardt RL*, Bando JK, Sullivan, BM, Locksley RM (2011) Divergent expression of IL-4 and IL-13 define unique functions in type 2 immunity. Nature immunology.13(1):58-66. (*equal contribution)
- Price AE, Liang HE, Sullivan BM, Reinhardt RL, Eisley CJ, Erle DJ, Locksley RM (2010) Systemically dispersed innate IL-13-expressing cells in type 2 immunity. Proc Natl Acad Sci USA.107(25):11489-94.
- Reinhardt RL, Liang HE, Locksley RM (2009) Cytokine-secreting follicular T cells shape the antibody repertoire. Nat Immunol 10(4): 385-393.
- Reinhardt RL*, Hong S*, Kang SJ, Wang ZE, Locksley RM (2006) Visualization of IL-12/23p40 in vivo reveals immunostimulatory dendritic cell migrants that promote Th1 differentiation. J Immunol 177(3): 1618-1627. (*equal contribution)
- Reinhardt RL, Bullard DC, Weaver CT, Jenkins MK (2003) Preferential accumulation of antigen-specific effector CD4 T cells at an antigen injection site involves CD62E-dependent migration but not local proliferation. J Exp Med 197(6): 751-762.
- Reinhardt RL, Khoruts A, Merica R, Zell T, Jenkins MK (2001) Visualizing the generation of memory CD4 T cells in the whole body. Nature 410(6824): 101-105.
Reviews:
- Dell’Aringa M and Reinhardt RL (2018) Using Cytokine Reporter mice to Visualize Type-2 Immunity in Vivo. Methods in Molecular Biology 1799 (1): 211-223
- Dell’Aringa M, Reinhardt RL, Friedman RS, Jacobelli, J. (2018) Live Imaging of IL-4-Expressing T Follicular Helper Cells in Explanted Lymph Nodes. Methods in Molecular Biology 1799 (1): 225-235
- Harmacek LD, Patel P, Woolaver R, Reinhardt RL, O’Connor BP (2018) Library Preparation for ATAC-Sequencing of Mouse CD4+ T Cells Isolated from the Lung and Lymph Nodes After Helminth Infection. Methods in Molecular Biology 1799 (1): 327-340
- Bao K, Reinhardt RL (2015) The differential expression of IL-4 and IL-13 and its impact on type-2 immunity. Cytokine 75(1):25-37
Complete List of Published Work in MyBibliography:
https://www.ncbi.nlm.nih.gov/sites/myncbi/richard.reinhardt.1/bibliography/48669013/public/?sort=date&direction=ascendingImmunology Microbiology
CU Anschutz
Research I North
12800 East 19th Avenue
Mail Stop 8333
Aurora, CO 80045
303-724-4224
School of Medicine
General
Students
CMS Login