Shared Content Block:
Styles for CDB Faculty Pages
Linda Barlow, Ph.D.
Professor

| [email protected] | |
| 303-724-3438 | |
| Ph.D., University of Washington - Seattle, 1991 | |
| Rocky Mountain Taste and Smell Center |
We rely upon our sense of taste to discern nutritious and delicious from spoiled food or even potentially toxic items. Taste buds are composed of a heterogeneous collection of sensory receptor cells, which transduce sweet, bitter, umami (savory), salt and sour into electrochemical signals that are transmitted by nerve fibers to the brain.
.jpg?sfvrsn=ae09bcba_2)
One particularly intriguing aspect of the taste system is that taste bud cells are continually renewed throughout life, in contrast to most of our peripheral sensory receptor cells such as rods and cones or hair cells of the inner ear, which lack this capability. Yet despite this regular turnover, our sense of taste is remarkably constant – sweet or salty foods eaten last year, when eaten today still taste sweet or salty. Intriguingly, taste function is often distorted in patients receiving targeted irradiation for head and neck tumors, as well as in cancer patients treated with a variety of specific chemotherapeutics, suggesting that taste cell turnover may be perturbed.
Finally, different people have different taste preferences, and this variability, in part, may reflect the number of taste buds found on the tongue. For example, “super tasters” appear to have an overabundance of taste buds, which occupy a large portion of the tongue surface, while most of us have taste buds distributed much less densely.
Thus, work in the lab focuses on 3 large questions, each of which is supported by a current NIH grant:
- How is the pattern, and therefore number, of taste buds established during embryonic development?
- Once established, how are taste bud cells continually renewed in adults?
- How do cancer therapies cause taste dysfunction?
We use both embryonic and adult mice to test hypotheses related to these main questions. Specifically, we employ drug-inducible, tissue-specific molecular genetics in mice to map the fate of subsets of cells in embryos and adults, as well as to delete or augment gene function in targeted cell populations. We evaluate the outcomes of these molecular genetic manipulations using a combination of immunofluorescence and in situ hybridization with antisense RNA probes assessed via conventional fluorescence microscopy, or spinning disc and laser scanning confocal microscopy.
We also employ quantitative PCR to determine how gene expression levels are altered in induced mutants compared with genetic controls. Additionally, we are gaining expertise in the powerful MATLAB platform to pursue more rigorous and automated quantitative analyses. Most recently, we have developed protocols for FACS to separate taste bud generating epithelial cells from non-sensory tongue epithelium, with the short term goal to use RNAseq to define and compare the transcriptomes of these two populations.
We also employ injury models to understand how the adult taste receptor cells regenerate, and gain insight into how taste function is perturbed in cancer patients. Toward this end, we have developed a mouse model of head and neck irradiation (Nguyen, Reyland and Barlow, 2012. J Neurosci), and are piloting similar studies using chemotherapeutics.
View Dr. Barlow's Latest Publications in PubMed
How are taste bud cells continually renewed in adults?
Castillo, D., K. Seidel, E. Salcedo, C. Ahn, F. De Sauvage, O.D. Klein and L.A. Barlow. 2014. Induction of ectopic taste buds by SHH reveals competency and plasticity of adult lingual epithelium. Development. 141(15): posted ahead of print July 3, 2014, doi:10.1242/dev.107631 Highlighted in the “In this Issue” section of the journal.
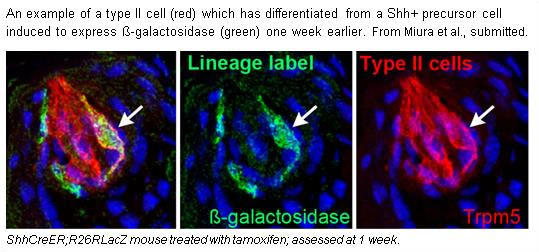
Miura, H., J.K. Scott, S. Harada and L.A. Barlow. 2014. Sonic hedgehog-expressing basal cells are general postmitotic precursors of functional taste receptor cells. Dev Dynamics. Mar 2. doi: 10.1002/dvdy.24121. [Epub ahead of print] PMID: 24590958
Gaillard, D. and L.A. Barlow. 2011. Taste bud cells in adult mice are responsive to Wnt/β-catenin signaling: implications for the renewal of mature taste cells. Genesis. 49(4):295-306. Article first published online 15 FEB 2011. PMC3536498
Miura, H. and Barlow, L.A. 2010. Taste bud regeneration, and the search for taste progenitor cells. Archives Italiennes de Biologie. 148:107-118. PMC3545678
Nguyen, H.M. and L.A. Barlow. 2010. BMP4 transgene expression differs in anterior fungiform versus posterior circumvallate taste buds of mice. BMC Neurosci. 11:129. PMC2966460
Seta, Y., C.L. Stoick-Cooper, T. Toyono, S. Kataoka, K. Toyoshima and L.A. Barlow. 2006. The bHLH transcription factors, Hes6, and Mash1, are expressed in distinct subsets of cells within adult taste buds. Arch Histol. Cytol. 69(3):189-198.
Seta, Y., C. Seta and L.A. Barlow. 2003. Notch-associated gene expression in embryonic and adult taste papillae and taste buds suggests a role in taste cell lineage decisions. J Comp Neurol 464:49-61.
How do cancer therapies cause taste dysfunction?
Nguyen, H.M., M.E. Reyland and L.A. Barlow. 2012. Mechanisms of taste bud cell loss following head and neck irradiation. Journal of Neuroscience. 32:3474-3484. PMC3320161
How is taste bud pattern established during embryonic development?
Kapsimali, M. and Barlow, L.A., 2013. Developing a Sense of Taste. Semin Cell Dev Biol. 24(3):200-9. PMC3604069 [Available on 2014/3/1]
Peterson, C.I, A.H. Jheon, P. Mostiwfi, C. Charles, S. Ching, S. Thirumangalathu, L.A. Barlow and O.D. Klein. 2011. FGF signaling regulates the number of posterior taste papillae by controlling progenitor field size. PLoS Genet 7(6): e1002098. Article first published online 02 JUNE 2011. PMC3107195
Harlow, D.E., H. Yang, T. Williams and L.A. Barlow. 2011. Epibranchial placode-derived neurons produce BDNF required for early sensory neuron development. Dev. Dynam. 240(2):309-23. Selected for "Highlights in DD". Selected by Faculty of 1000 Biology http://f1000.com/11607956 PMC3070660
Thirumangalathu, S., D.E. Harlow, A.L. Driskell, R.F. Krimm, and L.A. Barlow. 2009. Fate mapping of mammalian embryonic taste bud progenitors. Development. 136(9):1519-28. PMC2674259
Krimm, R.F. and Barlow, L.A. 2008. Development of the Taste System. In: A.I. Basbaum, A. Kaneko, G.M. Shepherd and G Westheimer, eds. The Senses: A Comprehensive Reference, Vol 4, Olfaction and Taste. S. Firestein and G.K. Beauchamp, San Diego: Academic Press, p.157-182.
Harlow, D.E. and L.A. Barlow. 2007. Epibranchial placodes give rise to sensory neurons which innervate taste buds. Dev. Biol. 310:317-328. Selected by Faculty of 1000 Biology.
Liu, F., S.Thirumangalathu, N.M. Gallant, C.L. Stoick-Cooper, S.H. Yang, S.T. Reddy, T. Andl, M.M. Taketo, A.A. Dlugosz, R.T. Moon, L.A. Barlow* and S.E. Millar*. 2007. Wnt-ß-catenin signaling initiates taste papilla development. *co-corresponding authors. Nature Genetics. 39:106-12.
Parker, M.A., M. Bell and L.A. Barlow. 2004. Cell contact-dependent mechanisms specify taste bud number and size during a critical period early in embryonic development. Dev. Dynamics 230:630-642.
Gross, J.B, A. Gottlieb and L.A. Barlow. 2003. Gustatory neurons derived from epibranchial placodes are attracted to, and trophically supported by taste bud-bearing endoderm in vitro. Dev Biol. 264:467-481.
Barlow, L.A. 2001. Specification of pharyngeal endoderm is dependent on early signals from axial mesoderm. Development 128(2): 4573-4583.
Barlow, L.A. Taste buds in ectoderm are induced by endoderm: Implications for mechanisms governing taste bud development. 2000. In: Regulatory Processes in Development: The Legacy of Sven Hörstadius. Proceedings of the Wenner-Gren International Symposium, L. Olsson and C.-O. Jacobson, eds. Portland Press, pp. 185-190.
Barlow, L.A. and R.G. Northcutt. 1997. Taste buds develop autonomously from endoderm without induction by cephalic neural crest or paraxial mesoderm. Development 124.05: 949-957.
Barlow, L.A., C.-B. Chien and R.G. Northcutt. 1996. Embryonic taste buds develop in the absence of innervation. Development 122(4): 1103-1111.
Barlow, L.A. and R.G. Northcutt. 1995. Embryonic origin of amphibian taste buds. Dev Biol 169: 273-285.