Shared Content Block:
Styles for CDB Faculty Pages
Yunsik Kang, Ph.D.
Assistant Professor

| yunsik.kang@cuanschutz.edu | |
| (303) 724.6475 | |
| Ph.D. University of Wisconsin Madison, 2016 | |
| Kang Lab Website |
Graduate Program Affiliations:
Cell Biology of Glial Phagocytosis
Our nervous system’s development is a gradual process that starts with an overabundance of neuronal connections and neurons. Just like an artist sculpts by carving out or removing excess material, the brain follows a similar process of refinement. This involves eliminating excess neurons and pruning unnecessary neuronal connections to create optimized neuronal circuits in the mature brain.
While it is well known that neurons and glia (the other main cell type in the nervous system) work together to refine these circuits, the exact molecular mechanisms behind this cooperation are not fully understood. Identifying these mechanisms is essential not only for understanding how the brain is built but also because defects in these processes are linked to neurodevelopmental disorders such as autism and schizophrenia.
In the Kang lab, we investigate how neurons and glia contribute to the refinement of neuronal circuits during development. To gain a comprehensive understanding of this process, we use the Drosophila (fruit fly) developing nervous system, which undergoes massive changes during metamorphosis. Our goal is to uncover the molecular mechanisms of neuronal remodeling and understand how abnormalities in this process are linked to neurodevelopmental disorders.
In addition to understanding nervous system development, studying the process of neuronal remodeling also provides insights into remarkable cell biology. Glia transform from cells that support neuronal function to ones that eat neurons, remodeling the entire nervous system within hours. How do these cells undergo such a dramatic transformation, and how do they ingest so much in such a short time?
One challenge with this level of consumption is producing enough membrane to keep forming phagosomes to degrade material while maintaining sufficient plasma membrane. Recently, we discovered that a bridge-like lipid transfer protein is crucial for this massive membrane expansion. We are using cell biology, genetics, and biochemical approaches to study the biology, function, and regulation of these fascinating proteins. Our lab will continue to explore these proteins in depth, along with other genes identified in screens for regulators of this process, to gain more new insights into this cell biology.
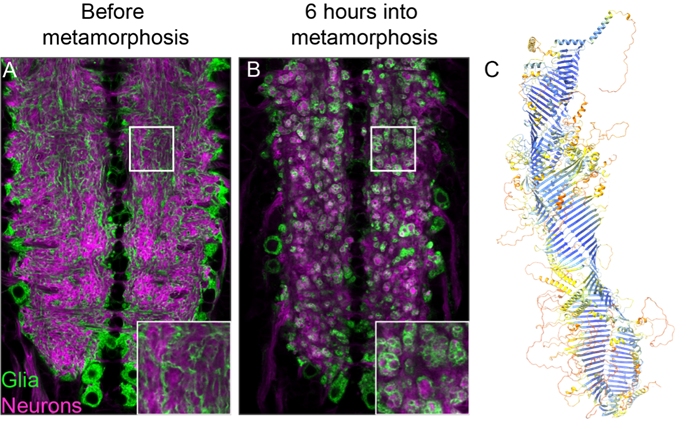
Kang Y, Lehmann K, Vanegas J, Long H, Jefferson A, Freeman M, Clark S. (2025). Structural basis of bulk lipid transfer by bridge-like lipid transfer protein LPD-3. Nature
Kang Y, Clark S. (2025). A protein tunnel that shuttles lipids around the cell. Nature Briefing
Lehmann K, Hupp M, Abalde-Atristain L, Cheng Y, Jefferson A, Sheehan A, Kang Y**, Freeman M**. (2025). Astrocyte-dependent local neurite Beat-Va neurons. Journal of Cell Biology(JCB)
**Co-corresponding author
Taylor J, Kang Y, Ouellet-Massicotte V, Kristine Micael M, Lacuros-Perkins V, Chen J, Sheehan A, Freeman M(2024). An in vivo genetic screen identifies Crq as a potent glial regulator of synapse elimination in development and aging. Under revision after review in Neuron.
https://www.biorxiv.org/content/10.1101/2024.06.24.600214v1
Chen J, Stork T, Kang Y, Sheehan A, Paton C, Monk K, & Freeman R. (2024). Astrocyte growth during morphogenesis is driven by the Tre1/S1pr1 phospholipid-binding G protein-coupled receptor Neuron,
Kang Y, JeffersonA, SheehanA, De La Torre R, Jay T, ChiaoL, Hulegaard A, Corty M, Baconguis I, ZhouZ, FreemanM(2023).Tweek-dependent formation of ER-PM contact sites enables astrocyte phagocytic function and remodeling of neurons. Under revision after review in Neuron.
https://www.biorxiv.org/content/10.1101/2023.11.06.565932v1
Clark S, Jeong H, Goehring A, Kang Y, Gouaux E. (2023). Large-scale growth of C. elegans and isolation of membrane protein complexes. NatureProtocols.
Jay T*, Kang Y*, Jefferson A, Freeman M. (2021). An ELISA-based method for rapid genetic screens in Drosophila. Proceedings of the National Academy of Sciences of the United States of America(PNAS).
*Contributed equally.
Hsu J, Kang Y, Corty M, Mathieson D, Peters O, Freeman M.(2021).Injury-Induced Inhibition of Bystander Neurons Requires dSarm and Signaling from Glia. Neuron.
Kang Y, Neuman S, Bashirullah A(2017). Tango7 regulates cortical activity of caspases during reaper-triggered changes in tissue elasticity. Nature Communications.
Kang, Y*, Marischuk K*, Castelvecchi G, Bashirullah A. (2017). HDAC Inhibitors Disrupt Programmed Resistance to Apoptosis During Drosophila Development. Genes, Genomes, Genetics(G3).
*Contributed equally.
Kang Y, Bashirullah A.(2014).A steroid-controlled global switch in sensitivity to apoptosis during Drosophila development. DevelopmentalBiology.