Shared Content Block:
Styles -- retool bulleted and numbered lists, add "spaced-out" class
RBI Informatics Fellows Program
Overview
Many facets of modern biological research involve the analysis of RNA to understand how cells control gene expression. These experiments involve powerful new sequencing methods that produce large data sets and require sophisticated bioinformatic analysis.
In the spring of 2016, faculty at the University of Colorado Anschutz Medical Campus were awarded $20 million to launch the RNA Bioscience Initiative (RBI). As one of its first acts, the RBI established the Informatics Fellows Program to recruit bioinformatics post-docs who would develop and apply computational approaches to RNA biology. In addition, the RBI Informatics Fellows have been tasked with training RNA biologists to use these methods in their own research, thereby alleviating the current bottleneck in analysis created by these large sequencing data sets.
A unique feature of this program is that RBI Informatics Fellows are themselves card-carrying RNA biologists who have significant training at the lab bench. This background gives fellows a rare perspective on RNA-related research and has been instrumental in helping collaborators ask and answer fundamental questions in RNA biology.
It’s an exciting time to be an RNA biologist capable of analyzing large sequencing data sets. We discover something new every day in our collaborators’ data sets and are capitalizing on our collective capabilities to push the limits of RNA sequencing technologies.
Objective
The objective of the RBI Informatics Fellows Program is to train the next generation of bioinformatic analysts. By the conclusion of their fellowship, these newly minted bioinformaticians will have mastered the skills, from both RNA biology and computer science, required to complete RNA sequencing experiments including design, execution, and analysis.
Activities
RBI Informatics Fellows divide their time between bioinformatic collaborations with other groups on campus, internal projects involving software and method development, and teaching.
Their training involves several important areas of bioinformatic collaboration:
- Experimental design. RNA sequencing experiments are easily conceptualized but require appropriate design to ensure rigorous analysis. Design considerations include experimental variables, e.g., replicates and conditions, sample preparation, e.g., RNA type and source, and the sequencing platform, e.g., short or long reads.
- Analytical software development. Once analyses are codified, we have worked to create and disseminate software for broad use in the community. Software development combines fundamental design principles with coding skills in Python, R, and C/C++ . Software developed in the program is available on our GITHUB page.
- Exploratory data analysis. RNA sequencing experiments often yield more questions than answers, so exploratory analyses that combine data mining and statistics are essential for a thorough investigation; our fellows’ biology training facilitates identification of meaningful signals in complex data sets. Deliverables are often fully reproducible dynamic documents that facilitate a collaborator’s own data exploration.
- Education and Outreach. Our educational mission is to broadly disseminate the analytical skills required to analyze RNA sequencing experiments– Fellows participate in various instructional activities, including hosting RBI Office Hours, mentoring RBI Summer Interns, designing and teaching RBI Workshops, and serving as teaching assistants for informatics Graduate Courses. These efforts have been impactful across campus, with many students, PRAs, postdocs, and principal investigators benefiting from the Fellows’ expertise.
It was the RBI Informatics Fellows Program that made me want to come to CU Anschutz. This program is an innovative way of facilitating collaborations between “wet labs” and bioinformaticians. The collaborations have been interesting and effective, pushing the RBI to be early adopters of novel RNA technologies– a great way to put CU Anschutz at the forefront of transformative science.
.jpg?sfvrsn=d0678db9_0)
Tyler Matheny
PhDMeet the Informatics Fellows
Group Leaders
- Neel Mukherjee, Ph.D. is a faculty advisor for the Informatics Fellows. The Mukherjee lab employs quantitative, transcriptome-wide experimental techniques and computational methods to investigate RNA regulatory networks in the context of human steroidogenesis and cancer.
- Jay Hesselberth, Ph.D. is co-director of the RBI and a faculty advisor for the Informatics Fellows program. The Hesselberth lab studies RNA damage and mechanisms of RNA repair and develops new molecular and computational approaches to advance single-cell analysis methods.
Current Fellows
- Ryan Sheridan, Ph.D. did thesis work at the University of Colorado Anschutz Medical Campus with David Bentley studying the role of backtracking in RNA Polymerase II proofreading. Ryan has been in the program since March 2019.
- Laura White, Ph.D. did thesis work at the University of Colorado Anschutz Medical Campus with Jay Hesselberth studying RNA repair mechanisms and developing novel repair detection methods. Laura has been in the program since August 2022.
- Christina Akirtava, PhD. did thesis work at Carnegie Mellon University with Joel McManus studying mechanism of translational initiation. Christina has been in the program since August 2023.
Former Fellows
- Kent Riemondy, Ph.D. is the research faculty lead of the Informatics Fellows. Kent conducts bioinformatics analysis of single cell sequencing datasets and develops software tools for analyzing RNA-Seq datasets. Kent was an RBI informatics fellow for 3+ years prior to transitioning to a research instructor.
- Michael Kaufman, Ph.D. did thesis work at the University of Colorado Anschutz Medical Campus with Joe Brzezinski studying the role of the transcription factor Otx2 in mouse retinal development. Michael has been in the program since November 2021.
- Tyler Matheny, Ph.D. did thesis work at the University of Colorado Boulder with Roy Parker studying the impact of cytoplasmic bodies on mRNA processing. Tyler was an RBI informatics fellow for 3 years.
- Kristen Wells-Wrasman, Ph.D. did thesis work at Stanford University with Lars Steinmetz studying immune cell development in the thymus and germinal centers. Kristen was in the program for 2 years. She is currently a Research Assistant Professor in the Barbara Davis Center at the University of Colorado Anschutz Medical Campus and works closely with the Sussel lab.
- Caitlin Winkler, Ph.D. did thesis work at the University of Colorado Anschutz Medical Campus with Santos Franco studying the role of Asc1 in development of cortical neurons. Caitlin was in the program for 2.5 years and is currently a Data Scientist at Bridge Informatics.
- Rui Fu, Ph.D. did thesis work at the University of California San Diego with Jens Lykke-Andersen studying the mechanism of nonsense-mediated mRNA decay. Rui was in the program for 4 years and is currently a Bioinformatics Scientist at the New York Genome Center.
- Austin Gillen, Ph.D. did thesis work at Northwestern University with Ann Harris studying miRNA regulation of CFTR. Austin was in the program for 3 years. Austin recently received a career development award from the Department of Veterans Affairs and works closely with the lab of Craig Jordan to understand heterogeneity in AML tumors.
Projects
The success of the Informatics Program can be measured in part by its impact on our collaborators’ publications and grants. Since the Program’s inception in 2016, the Informatics Fellows have:
- Initiated 186 projects among 122 unique investigators
- Completed 130 projects, leading to a total of 47 publications, with another 7 actively in preparation VIEW PUBLICATIONS
- Contributed analyses to multiple NIH, NSF and other grant applications from investigators across campus
In 2017 the RBI purchased a Chromium Controller from 10x Genomics to facilitate the construction of single-cell mRNA sequencing libraries (this instrument is maintained in the Genomics Core). Due to the success of this method, 103 experiments to-date have focused on single-cell mRNA sequencing. Figure 1 below details these scRNA-sequencing experiments, as well as a wide variety of other RNA-related projects.
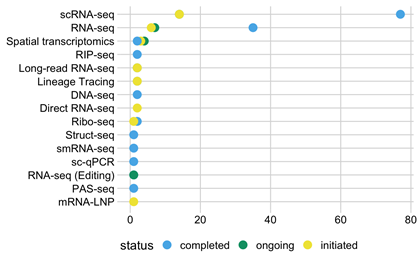
Figure 1: Project types and status.
RBI-Funded Projects
Through its Pilot Grant Program, the RBI has funded more than 119 RNA-focused projects, catalyzing interest in and adoption of cutting-edge technologies among campus investigators. Funded projects are designed, executed, and analyzed by the RBI Informatics Fellows. In addition to the projects funded through the RBI Pilot Grant Program, the RBI has provided both experimental and bioinformatic support via several mechanisms:
- Single-cell mRNA sequencing demonstrations and platform trials- To enhance campus capabilities in RNA sequencing, the RBI has invited several companies (Wafergen, Illumina, 10x Genomics) to demonstrate their system capabilities on campus.
- Collaborative Projects with RBI Faculty Hires- Data generated by the Informatics Fellows will be instrumental in helping these new faculty publish original research and obtain external funding.
- Other Collaborations- The RBI has actively sought collaborations with campus investigators, including clinicians. We invite you to contact Jay Hesselberth ([email protected]) for more information.
Figure 2 summarizes the 161 RBI-funded projects analyzed by the Informatics Fellows to date.
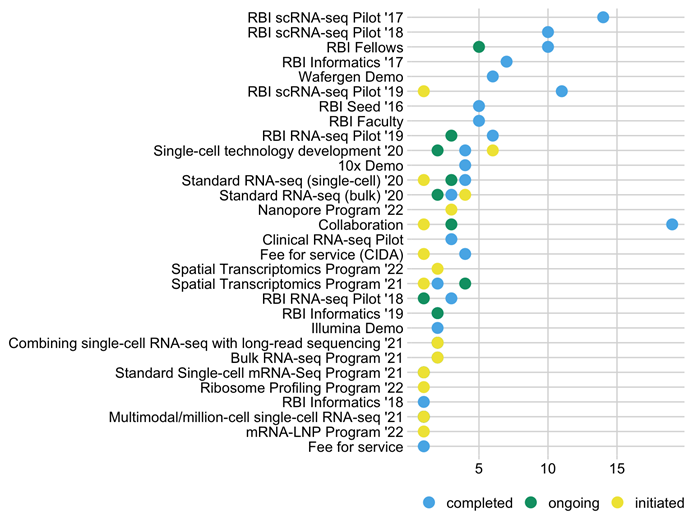
Figure 2: Project source organized by RFA.
Fellow-Initiated Projects
In collaboration with the Hesselberth Lab, the RBI Informatics Fellows also work on their own projects. These include software development, e.g., R packages for genome interval analysis and for the analysis of single-cell functional heterogeneity, new molecular methods focused on single-cell RNA-seq, and bioinformatics curriculum development.
| Fellow-Initiated Internal Projects | ||
|---|---|---|
| Description | Type | Status |
| single cell PAS-Seq analysis | Method Development | ongoing |
| R Package development (djvdj) | Method Development | ongoing |
| R Package development (raer) | Method Development | ongoing |
| Targeted single cell RNA-Seq resampling | Method Development | completed |
| R Package development (valr) | Method Development | completed |
| R Package development (scrunchy) | Method Development | completed |
| single cell classification from bulk RNA-Seq | Method Development | completed |
| RBI Office Hours | Teaching | ongoing |
| scRNA-seq short course | Teaching | ongoing |
| Practical Biological Data Analysis in R | Teaching | completed |
| Genome Analysis Workshop | Teaching | completed |
| RNA secondary structure short course | Teaching | completed |
| Mentoring- RBI interns, students, fellows | Teaching | completed |
Other Outreach
The RBI can also provide letters of support to investigators for their grant applications that detail the Informatics Fellows program and the broader RNA sequencing and other single cell analytic capabilities on campus. Please contact Jay Hesselberth ([email protected]) or details.
The RBI Informatics Fellows also host office hours to provide help with programming, software, pipelines, and experimental design questions. Please visit the RBI Office Hours page for scheduling.