Shared Content Block:
Styles -- retool bulleted and numbered lists, add "spaced-out" class
Pilot Grant Program
APPLICATIONS FOR THE FALL 2023 RBI PILOT GRANT PROGRAM ARE CLOSED!
Program Statistics 2016-2021
120 RBI Pilot Grants Awarded
- 67 projects completed
- 28 projects are still in progress/have not yet started
- 4 grants were abandoned due to insurmountable technical difficulties
- 21 awardees yet to report the outcomes of their award(s)
103 Investigators Participated
- 3 AMC Schools- School of Medicine, School of Dental Medicine, Skaggs School of Pharmacy
- 16 Departments
- 8 Medicine Divisions
- 5 Pediatric Divisions
- 2 Surgery Divisions
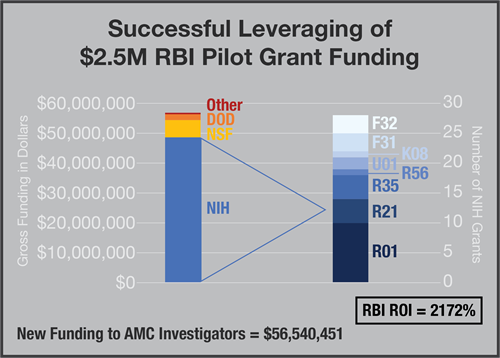
Total Funds Spent by RBI = $2,488,631
Gross Return by Investigators = $56,540,451
Net Return to AMC = $54,051,820
Fold Increase = 21.72
Return On Investment (ROI) = 2172%
Thanks for the support! It truly was “transformative”. Data from our 2017 RBI Pilot Grant project supported 2 publications and an NIH R35 for $6,904,527 in total costs.

Bruce Appel, PhD
Section Head, Professor of Pediatrics, Diane G. Wallach Chair of Pediatric Stem Cell BiologyShared Content Block:
Styles -- modify testimonial colors
Program Impacts 2016-2021
Financial-
- 40 new grants funded using data/findings derived from RBI Pilot Grants (28 NIH, 2 NSF, 1 DOD, 9 Other)
- 6 R01’s PENDING with fundable scores
- 16 additional grants have been submitted (NIH R01, NIH R21, NSF, DOD, Cancer Center, societies/foundations)
- 15 grants in preparation
Visibility-
- 34 publications in 27 different journals
- 7 submitted manuscripts
- 31 manuscripts in preparation
- 3 patent applications
- Collaborations with CU Boulder, Arizona State, University of Tennessee, University or Iowa, and others
RBI Pilot Grant Awardees
RECIPIENT | DEPARTMENT | AWARD | PROJECT |
Santos Franco, PhD | Pediatrics- Developmental Biology | Spatial Transcriptomics | Elucidating the spatiotemporal mechanisms of neural stem cell fate specification in the developing brain |
Katharina Hopp, PhD | Medicine- Renal Diseases and Hypertension | Spatial Transcriptomics | Utilizing spatial transcriptomic profiling to understand the heterogenous histopathology of polycystic kidney disease |
| Eric Clambey, PhD | Immunology & Microbiology | mRNA-LNP | mRNA-LNP vaccination against chronic gammaherpesvirus infection |
| Heide Ford, PhD | Pharmacology | Ribosome Profiling | Determining the Mechanism of Action of 8430, a Novel Inhibitor of Metastasis |
| Matthew Gailbraith, PhD | Pharmacology | Nanopore | Long-read RNA-sequencing to discover novel transcriptome features in Down syndrome |
| Jeffrey Kieft, PhD | BMG | Nanopore | Using long-read NGS to interrogate flavivirus noncoding RNA structure and modifications |
| Matthew Taliaferro, PhD | BMG | Nanopore | Nanopore-based detection of 8-oxoguanosine for RNA localization analysis |
RECIPIENT | DEPARTMENT | AWARD | PROJECT |
David Bentley | BMG | RNA-seq | Determination of nascent RNA folding in vivo by psoralen cross-linking |
James Maloney | Medicine- Pulmonary Sciences and Critical Care | RNA-seq | Gene expression changes in human T-regs in response to the SARS-CoV-2 nsp15 protein |
Tania Reis | Medicine- Endocrinology, Metabolism, and Diabetes | RNA-seq | Dissecting m6A's role in metabolism |
Chandra Tucker | Pharmacology | RNA-seq | Probing RNA transcriptional responses to sudden appearance or dissolution of biomolecular condensates |
Ethan Hughes | CDB | scRNA-seq | Determine the transcriptomics profile of mature oligodendrocytes participating in remyelination |
Elena Hsieh | Pediatrics-Allergy/Immunology | scRNA-seq | Understanding SARS-CoV-2 B cell responses derived from COVID-19 immunization |
Suja Jagannathan | BMG | scRNA-seq | Singe cell long-read CaptureSeq to characterize isoform structure of DUX4 transcripts |
Matt Kennedy | Pharmacology | scRNA-seq | Profiling amyloid-beta binding cells of the central nervous system |
Tem Morrison | Immunology and Microbiology | scRNA-seq | Mechanisms of chronic chikungunya virus infection and joint disease |
Holger Russ | BDC; Immunology and Microbiology | scRNA-seq | Cellular heterogeneity and function in thymi of patients with Down syndrome |
Charles Sagerstrom | Pediatrics-Developmental Biology | scRNA-seq | Adaptation of single-cell calling card technology to test TF function in zebrafish embryogenesis |
Rebecca Schweppe | Medicine- Endocrinology, Metabolism, and Diabetes | scRNA-seq | Deregulation of the transcriptome promotes the transformation of anaplastic thyroid cancer |
Lori Sussel | BDC | scRNA-seq | Identification of pancreatic islet cell-type specific splicing events |
Benjamin Bitler | OBGYN | Spatial Transcriptomics | Spatial analysis of the ovarian cancer tumor microenvironment |
Nicholas Foreman | Pediatrics-Hematology, Oncology, and Bone Marrow Transplantation | Spatial Transcriptomics | Spatial lineage trajectory analysis in childhood brain tumor medulloblastoma |
Wendy Macklin | CDB | Spatial Transcriptomics | Spatiotemporal analysis of microglial contribution to immune-mediated demyelinating lesion formation |
Rebecca McCullough | Pharmaceutical Sciences | Spatial Transcriptomics | The neuroimmune landscape of the aging brain: Dissecting the impact of alcohol |
Julie Siegenthaler | Pediatrics- Developmental Biology | Spatial Transcriptomics | Spatial transcriptomics to study mechanisms of meninges development |
Carmen Sucharov | Medicine- Cardiology | Spatial Transcriptomics | Spatial transcriptomics in the failing pediatric heart |
Kelly Sullivan | Pediatrics- Developmental Biology | Spatial Transcriptomics | Defining the impact of interferon receptor copy number on neuroinflammation in a mouse model of Alzheimer’s disease in Down syndrome |
RECIPIENT | DEPARTMENT | AWARD | PROJECT |
Kristin Artinger | Craniofacial Biology | RNA-seq | Using RNA-seq to identify molecular interactions among different cell populations during neural crest cell migration |
Adela Cota-Gomez | Pulmonary Sciences and Critical Care | RNA-seq | Nrf2 activator PB125 as a potential therapeutic agent against coronavirus (COVID related) |
Tom Evans | CDB | RNA-seq | Discovering direct RNA targets of novel ncRNAs |
Wendy Macklin | CDB | RNA-seq | Developmental RNAseq analysis of the translatome in axons with conditional neuronal Raptor deletion |
Nidia Quillinan | Anesthesiology | RNA-seq | Distinct firing properties of thalamic neurons receiving cerebellar input |
Olivia Rissland | BMG | RNA-seq | COVID genomic sequencing and tracking |
Rebecca Schweppe | Pathology | RNA-seq | Nuclear FAK alters the transcriptome to promote thyroid cancer growth and survival |
Linda van Dyk | Barbara Davis Center | RNA-seq | Identification of host and viral ncRNA interatcing with La and RIF-I during gammaherpesvirus infection |
Natalia (Maria) Vergara | Ophthalmology | RNA-seq | Unraveling the molecular basis of retinal abnormalities in Down syndrome |
Quentin Vicens | BMG | RNA-seq | Using RNA-seq to determine the biological role of the Z-RNA binding domain of ADAR1 during the interferon response |
Rocky Baker | Immunology and Microbiology | scRNA-seq | Combined TCR and gene expression profile of HIP-reactive T cells in islet grafts after induction of tolerance with 2.5HIP PLG-NPs |
David Beckham | Infectious Disease | scRNA-seq | Defining innate immune responses to SARS-CoV2 with single cell resolution |
Melanie Koenigshoff | Pulmonary Sciences and Critical Care | scRNA-seq | Unraveling the molecular mechanisms of distal lung epithelial stem cell impairment in COPD |
Liyen Loh | Immunology and Microbiology | scRNA-seq | Deciphering the development of innate T cells in the human thymus |
James Nichols | Craniofacial Biology | scRNA-seq | Clonal dominance in appendage development |
Matthew Sikora | Pathology | scRNA-seq | Functional heterogeneity in estrogen receptor co-regulator activity |
Mary Weiser-Evans | Renal Diseases and Hypertension | scRNA-seq | SMC-derived AdvSca-SM cell contribution to cardiac fibrosis |
Timothy McKinsey | Cardiology | scRNA-seq | Exploring the therapeutic potential of HDAC inhibition in HFpEF |
Howard Davidson | Barbara Davis Center | scTech Development | Monitoring alternative splicing in single human T cells by ScISOr-Seq plus |
Peter Dempsey | Gastroenterology, Hepatology, and Nutrition | scTech Development | Population-based analysis of intestinal organoid heterogeneity using CellTagging and scRNAseq |
James Dylewski | Renal Diseases and Hypertension | scTech Development | The role of FcRn in antigen presentation by glomerular endothelial cells in immune mediated kidney disease |
Rachel Friedman | Barbara Davis Center | scTech Development | Myeloid cells in the islets |
Kirk Hansen | BMG | scTech Development | Development of an ECM atlas to guide regenerative medicine therapies using MULTI-seq |
Elena Hsieh | Pediatric Allergy and Immunology | scTech Development | Understanding IL-15/IL-2RB interactions in a hypomorphic IL-2RB mouse model |
Katja Kiseljak-Vassiliades | Endocrinology, Metabolism, and Diabetes | scTech Development | In situ reconstruction of distinct cell populations within the human adrenal gland |
Christian Mosimann | Pediatrics | scTech Development | Charting cardiovascular progenitor clonality and developmental trajectories in toto |
Eric Pietras | Hematology | scTech Development | Clonal tracking and expression analysis in a mouse model of human clonal hematopoiesis of indeterminate potential (CHIP) |
Stephen Santoro | Peds-Developmental Biology | scTech Development | Lineage tracing the odorant receptor fates of olfactory sensory neurons |
Eszter Vladar | Pulmonary Sciences | scTech Development | WNT4 signaling specifies ciliated cell fate in the airway epithelium |
Tuoqi Wu | Immunology and Microbiology | scTech Development | Identify the transcriptional and protein expression signatures of CAR T cells with long-term antitumor immunity using endogenous TCR sequences as molecular barcodes |
RECIPIENT | DEPARTMENT | AWARD | PROJECT |
Howard Davidson | Barbara Davis Center | Bioinformatics Support | Using CITE-seq to define immune biomarkers of the honeymoon period in type 1 diabetes |
Joshua Black | Pharmacology | RNA Biology | Enrichment detection and discovery of repeat element-gene mRNA fusion transcripts |
Tom Evans | Cell and Developmental Biology | RNA Biology | Control of RNP co-assembly dynamics in early development |
Tania Reis | Endocrinology, Metabolism, and Diabetes | RNA Biology | Mechanisms of control of Drosophila fat storage by conserved RNA-binding protein Nito |
Douglas Shepherd & Diego Restrepo | Pharmacology | RNA Biology | High-throughput spatial quantification of RNA expression in mouse olfaction after exposure to viral agents |
Jianbin Wang | BMG | RNA Biology | Transcription and transcription-replication collision and R-loop formationduring programmed DNA elimination |
Diana Cittelly | Pathology | RNA-seq | Transcriptional profile of reactive astrocytes at early and late stages of brain metastatic progression |
Katherine Fantauzzo | Craniofacial Biology | RNA-seq | Srsf3-mediated alternative RNA splicing downstream of PDGFRalpha signaling in the palatal mesenchyme |
Jeffrey Kieft | BMG | RNA-seq | Identification of cellular exoribonuclease-resistant RNAs by xrRNA-seq |
Traci Lyons | Oncology | RNA-seq | Investigating SEMA7A-regulated mechanisms of endocrine resistance in ER+ breast cancer |
Michael McMurray | CDB | RNA-seq | Defining the maternal contribution of RNAs in yeast gametes |
Chad Pearson | CDB | RNA-seq | Identifying centriolar RNAs in Tetrahymena thermophila |
Rytis Prekeris | CDB | RNA-seq | Identification of midbody-associated RNA |
Eszter Vladar | Pulmonary Sciences | RNA-seq | Molecular, physiological and health outcome impacts of triple combination therapy in cystic fibrosis airway disease |
Trevor Williams | Craniofacial Biology | RNA-seq | Functional analysis of differential splicing by Rbm10 – the gene mutated in human TARP Syndrome |
Rocky Baker | Immunology and Microbiology | scRNA-seq | Monitor phenotype and TCR of HIP-reactive T cells over time in a subject at risk for T1D |
Joseph Brzezinski | Opthamology | scRNA-seq | Regulation of human cone photoreceptor development |
Eric Clambey | Anesthesiology | scRNA-seq | CD4 T cell lineage tracing by scRNA-seq |
Nicholas Foreman | Pediatrics | scRNA-seq | To investigate the feasibility of single-nuclei RNAseq analysis of pediatric brain tumors |
Dany Gaillard | CDB | scRNA-seq | Single cell RNA-seq of irradiated mouse CVP |
Ross Kedl | Immunology and Microbiology | scRNA-seq | In search of Tvacs: single cell RNAseq comparison between vaccine-elicited and infection elicited CD8+ T cell responses |
T. Rajendra Kumar | OB/GYN | scRNA-seq | Single cell RNA-seq analysis of pituitary gonadotropes |
Wendy Macklin | CDB | scRNA-seq | scRNAseq of oligodendrocytes during development with conditional rictor deletion in neurons |
Karen Moulton | Cardiology | scRNA-seq | Translating vascular cell phenotypes of human coronary artery disease |
Holger Russ | Barbara Davis Center | scRNA-seq | Elucidating AIRE dependent expression of tissue-restricted antigens in stem cell derived organotypic thymus cultures from healthy and autoimmune patients |
Daniel Sherbenou | Hematology | scRNA-seq | Tracking resistant multiple myeloma cells in residual disease by single-cell RNA-seq |
Tamara Terzian | Oncology | scRNA-seq | Molecular determinants of lymphangiogenesis |
RECIPIENT | DEPARTMENT | AWARD | PROJECT |
Lori Sussel | Barbara Davis Center | Bioinformatics Support | Identification of IncRNAs associated with obesity-induced diabetes |
Kristin Artinger & David Bentley | Craniofacial Biology & BMG | RNA-seq | Developmental control of co-transcriptional splicing in zebrafish |
Sue Kinnamon | Otolaryngology | RNA-seq | Transcriptome analysis of GAD65-expressing taste receptor cells |
Suchitra Rao | Infectious Diseases | RNA-seq | Measures of immune responses to influenza infection in children |
Lori Sussel | Barbara Davis Center | RNA-seq | Determine the effect of zinc deficiency on the islet transcriptome |
Yael Aschner | Pulmonary Sciences and Critical Care Medicine | scRNA-seq | Analysis of ARDS BAL |
Matthew Burchill | Gastroenterology and Hepatology | scRNA-seq | Dissection of the T lymphocyte activation landscape in NASH |
David Clouthier | Craniofacial Biology | scRNA-seq | A transcriptional code coordinates jaw development |
Howard Davidson | Barbara Davis Center | scRNA-seq | Profiling T cell responses to low affinity polyclonal stimuli |
Jonathan Kurche & David Schwartz | Pulmonary and Critical Care | scRNA-seq | Single nuclei RNA sequencing in idiopathic pulmonary fibrosis (IPF) |
Hong Li and Eriv Van Otterloo | Craniofacial Biology | scRNA-seq | Redefining the ameloblast lineage |
Martin McCarter | Surgical Oncology | scRNA-seq | Analysis of tumor infiltrating MDSCs in human melanoma |
Joanna Poczobutt & Oliver Eickelberg | Pulmonary and Critical Care | scRNA-seq | The role of MZB1-positive plasma B cells in idiopathic pulmonary fibrosis |
Beth Tamburini | Immunology | scRNA-seq | Transcriptional profiling of lymphatic endothelial cells and dedritic cells during antigen archiving exchange |
Ivana Yang | Pulmonary and Critical Care | scRNA-seq | Characterization of bronchoalveolar lavage cell populations from patients with sarcoidosis |
RECIPIENT | DEPARTMENT | AWARD | PROJECT |
Steve Anderson | Pathology | Bioinformatics Support | Analysis of the role of glucose metabolism upon tumor-initiating cells |
David Bentley | BMG | Bioinformatics Support | The development of a bioinformatics pipeline to characterize nascent RNA structure |
Nicholas Foreman | Hematology | Bioinformatics Support | Single-cell mRNA seq analysis of pedatric brain tumors |
Bernice Gitomer | Renal Medicine | Bioinformatics Support | Identification of the etiology of the bone defect in patients with autosomal dominant polycystic kidney disease (ADPKD) |
Eva Grayck | Cardiovascular and Pulmonary | Bioinformatics Support | Bioinformatic analysis of RNA sequencing data to interrogate the redox regulation of alveolar macrophage populations in acute lung injury |
Craig Jordan | Hematology | Bioinformatics Support | Identification and characterization of drug-resistant subpopulations in AML |
Elizabeth Wellberg | Pathology | Bioinformatics Support | Using gene expression profiles to predict obesity-associated breast cancer progression |
Bruce Appel | Pediatrics, Stem Cell Biology | scRNA-seq | Single cell RNA-seq investigation of glial cell heterogeneity |
Kristin Artinger | Craniofacial Biology | scRNA-seq | Single cell profiling of neural crest derived cancers |
Linda Barlow | CDB | scRNA-seq | Single cell RNA-seq of Sox2-GFP lingual epithelial cells |
Dick Davis | BMG | scRNA-seq | Single cell RNA-seq analysis of ascarsis development |
Peter Dempsey | Gastroenterology, Hepatology, and Nutrition | scRNA-seq | Reversion of Defa4Cre-expressing Paneth cells to a stem cell state in response to intestinal injury |
Robert Doebele | Oncology | scRNA-seq | Single cell analysis of oncogene-driven lung adenocarcinoma to determine mechanisms of residual disease following oncogene-targeted therapy |
Patricia Ernst | Pharmacology | scRNA-seq | Enhancing hematopoietic stem cell emergence from pluripotent embryonic stem cells using MLL1 induction |
James Nichols | Craniofacial Biology | scRNA-seq | Understanding jaw evolution by comparing craniofacial progenitor cells in lamprey and zebrafish |
Holger Russ | Barbara Davis Center | scRNA-seq | Elucidating human beta cell subtype regulation and interconversion in health and disease |
Howard Davidson | Barbara Davis Center | scRNA-seq | Single cell profiling in type 1 diabetes |
Joan Hooper | CDB | scRNA-seq | Analysis of craniofacial development |
Lori Sussel | Barbara Davis Center | scRNA-seq | Analysis of pancreatic progenitors |
RECIPIENT | DEPARTMENT | AWARD | PROJECT |
Erin Baschal | Orthopedics | Clinical | Comparison of facet joint cartilage RNA from adolescent idiopathic scoliosis patients and control individuals |
Kathleen Connell | Urogynecology and Reconstructive Surgery | Clinical | Epigenetic and RNA expression profiles in women with pelvic organ prolapse |
James Costello | Pharmacology | Clinical | RNA sequencing analysis of bladder cancer cell lines that localize to the lung and liver |
Tom Evans | CDB | Seed Grant | Identification of novel RNA targets of sm-rings |
Bernice Gitomer | Renal Diseases and Hypertension | Seed Grant | Prediction of the rate of autosomal dominant polycystic kidney disease progression based on circulating miRNA levels |
Aaron Johnson | BMG | Seed Grant | m6A RNA modification in long noncoding RNA matchmaking and heterochromatin formation |
Sandy Martin | CDB | Seed Grant | Investigating RNA editing in the brain of mammalian hibernator and Investigating transcript isoform dynamics in a hibernator |
Lee Niswander | Molecular, Cellular & Developmental Biology | Seed Grant | An alternative splicing mechanism in the lincRNA-1354 locus commits embryonic stem cells to neural differentiation |
Eric Pietras | Hematology | Seed Grant | Role of RNA splicing factor U2AF1 mutations in regulating hematopoietic stem cell inflammation response and myelodysplastic syndrome |
Jennifer Richer | Pathology | Seed Grant | Targeting the long non-coding RNA MALAT1 in ovarian cancer |
Mario Santiago | Infectious Diseases | Seed Grant | Noncoding RNAs in mucosal HIV-1 persistence and pathogenesis |
Carol Sartorius & Peter Kabos | Pathology & Oncology | Seed Grant | Single cell analysis of organ specific metastasizing cells in estrogen receptor positive breast cancer |
Lori Sussel | Barbara Davis Center | Seed Grant | lncRNA discovery in motor neurons |
Trevor Williams | Craniofacial Biology | Seed Grant | Probing the consequences of the disruption of epigenetic bookmarking in human Branchio-oculo-facial Syndrome using single-cell sequencing |