Meet the Scientist
Dr. Catherine Musselman
Associate Professor
Department of Biochemistry and Molecular Genetics

Cat investigates chemical signals that influence how DNA is packaged and used inside our cells
Did you know that every single cell in our body has the same DNA inside but only uses the genes it needs to become the cell type that it is? It’s true! As an example, skin cells only use the genes needed to create all the proteins and components essential to becoming a skin cell. They leave all the other genes unused. The same is true for the cells in our hearts, lungs, eyes, etc. You name the cell type and it only uses the genes needed to become that specific type of cell while leaving the other genes unused.
How do our cells know which genes on our DNA to use?
Our DNA is kind of like a cook book that is filled with instructions (or recipes) to make thousands of proteins. Think about the classic cookbook, ‘The Joy of Cooking’ - it contains recipes for all different types of food, such as casseroles, cheeseburgers, cakes, and cookies. You might be interested in baking a birthday cake so you select and use a recipe for chocolate cake while leaving all of the other 100s of recipes unused. That’s what happens inside every cell in our body every day.
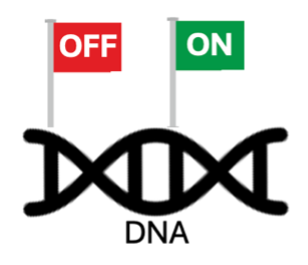
Dr. Catherine “Cat” Musselman is a research scientist in the Biochemistry and Molecular Genetics Department who studies how genes can be turned ‘ON’ or ‘OFF’ within a field of science known as ‘epigenetics.’ The term epigenetics means “on top of” or “above” the genome. Cat and her team study the chemical signals that are placed on top of genes to control how they’re used – they don’t actually change the sequence of the DNA just how it’s used. These chemical signals can be thought of as flags that are added to the outside of your DNA to label which genes need to be used and are turned ‘ON’ (think green flag) and which genes are not needed and should stay turned ‘OFF’ (think red flag).
Epigenetics - or signals on top of the genome - are kind of like the messages written in frosting on top of a cake. Some cakes have the message ‘happy birthday’ written on top while others display ‘congratulations’ or ‘happy anniversary.’ The messages written on a cake act as a signal for the event or the activity the cake was made for but won’t change the cake inside.
How are our genes stored?
Humans have around 20,000 genes that serve as instructions to make all of the proteins that are needed for our bodies to be built and work. These genes are written in the chemical language of DNA, and incredibly, all of our genes are written on long strands of DNA that measure 6 feet inside each cell. Yes, 6 feet of DNA inside each cell! You might imagine that it is quite a feat to carefully wrap up 6-feet of DNA carefully inside each cell so that it doesn’t turn into an impossibly tangled mess. To keep the DNA organized inside each cell, it is carefully wound around a group of proteins called histones. Histones look like the plastic center part of a yo-yo and the DNA is the string that wraps around the center groove. Wrapping the strands of DNA around the histone (yo-yo) core makes it look like ‘beads on a string’ and keeps it from becoming a tangled mess inside the cell.
Have you ever heard the saying, ‘opposites attract?’ This saying describes how histones and DNA work. Histones and DNA are attracted to one another due to their opposite chemical charges. Histones have an overall positive charge and DNA has an overall negative charge. This opposite attraction allows 6-feet of negatively charged DNA to be wound around positively charged histones inside each cell to create a tight and compact bundle of genetic instructions. Histones function to package a huge amount of DNA into a very small space and they also serve as signaling platforms (recall the green and red flags) to regulate access and use of the underlying genetic material.
Have you ever heard the saying, ‘opposites attract?’ This saying describes how histones and DNA work. Histones and DNA are attracted to one another due to their opposite chemical charges. Histones have an overall positive charge and DNA has an overall negative charge. This opposite attraction allows 6-feet of negatively charged DNA to be wound around positively charged histones inside each cell to create a tight and compact bundle of genetic instructions. Histones function to package a huge amount of DNA into a very small space and they also serve as signaling platforms (recall the green and red flags) to regulate access and use of the underlying genetic material.
How do our cells know which genes to turn on and off?
Modifications to histones through the addition of a variety of small chemical groups are called post-translational modifications (PTMs). Histone PTMs are the red and green flags mentioned above that alter access to DNA and play a role in turning genes ‘ON’ or ‘OFF.’ Histone PTMs are known to play a huge role in defining cell identity and allowing each cell to respond to the environment. This environment includes your diet, emotional stress, environmental toxins, and infection. Histone PTMs change in response to all of these factors and can also be messed up due to genetic mutation, including mutation of the histones themselves. Since histone PTMs are relatively easy to manipulate through changes to diet and drugs, they have high therapeutic potential. However, to understand how to best use histone PTMs to develop therapeutics that would allow us to change the way DNA is used, we must first understand how they are functioning.
What experiments are Cat and her research team doing to crack the histone code?
The Musselman lab created a model of how the histones are interacting with DNA and how histone PTMs may alter or change these interactions. Their next steps are to test this model more thoroughly by investigating how targeted changes in histone PTMs and/or histone mutations change gene expression. Some of these experiments have been done in partnership with a company called Epicypher focused on discovering and studying key parts of the histone code. The research in the Musselman is focused on cracking the histone code and learning more about how we can use that knowledge to develop drugs that treat human disease.
If you want to learn more about the scientist, please head to their official CU webpage.