Services
Microbiome Sequencing
In order to support characterization of microbial community composition and activity in mucosal biospecimens and tissues, we provide high-quality and quality-controlled microbial diversity data by performing nucleic acid isolation from biospecimens, 16S rRNA PCR and sequencing on a discounted fee-for-service basis to RDRRC members.
Microbiome Services are provided through the Anschutz Center for Microbiome Excellence (ACME), which provides microbiome services through the lab of Catherine Lozupone.
Nucleic acid isolation and 16S rRNA sequencing used the standardized protocols of the Earth Microbiome project described at https://earthmicrobiome.org/protocols-and-standards/.
We have provided quality 16S rRNA targeted sequencing data for biospecimens including feces, mucosal biopsy, cecum, and saliva for more than 25 research groups supporting more than 15 different publications.
Metabolomics
In order to support characterization of the metabolome of biospecimens and tissues of importance in rheumatic disease pathogenesis, we provide high-quality and quality-controlled untargeted and targeted metabolomics/lipidomics data on a discounted fee-for-service basis for RDRRC members. The Reisdorph Lab also offers four-day Hands-On Metabolomics Workshops and four-day Proteomics workshops approximately once per year. Participants will learn introductory metabolomics or proteomics science and applicable protocols and technologies through a comprehensive, hands-on exploration of typical mass spectrometry-based workflows.
The SSPPS Mass Spectrometry Facility will provide method development, and discounted services (Table 2) to support RDRRC Investigators and trainees in metabolomics and lipidomics studies. The facility maintains 14 mass spectrometers with capabilities spanning proteomics, metabolomics, targeted small molecule assays, and mass spectrometry imaging. In addition to the Director, Dr. Nichole Reisdorph, the MS core staff is comprised of 2 PhD scientists, 4 scientific staff members, and 1 administrator. Services include preparation of samples, including plasma, urine, feces, cells, and intestinal tissues, comprehensive metabolome and lipidome coverage, and data analysis. Targeted, quantitative assays, including quantitation of 94 bioactive lipid mediators, bile acids, amino acids, short chain fatty acids, steroids, nucleotides, endocannabinoids, neurotransmitters, FAMES, and acylcarnitines are also available. Consulting with staff is available to help tailor the experiments to suit specific needs. We can also help with initial experimental design, including assistance with sample collection and storage techniques to optimize downstream results.
For more information, email Richard Reisdorphat [email protected]

Data Analysis Support
In order to support data analysis to assess host:microbe relationships, we will provide pre and post study statistical design consultation and support for mucosal microbiome and integrated multi’omic analyses on a free/discounted fee-for-service basis for RDRRC members.
Pre study statistical design consultation:
Pre-sequencing consultation will include study design elements critical and specific to performing microbiome:host interaction research. For instance, for microbiome, this may include selecting appropriate controls given knowledge of factors that influence microbiome composition across healthy people. This will be done in collaboration with the Population and Data Sciences Core (Core 1), to ensure that appropriate epidemiology data, such as dietary intake, is obtained for a study. We also provide guidance and training on tools available for enabling the informed selection of cohort sizes. Initial study design consultation will be provided free-of-charge to RDRRC investigators to facilitate the initiation of well-designed research projects.
Microbiome data analysis support (16S):
We provide core 16S rRNA microbiome analyses for a standard fee (Table 2). These include demultiplexing and denoising sequences and producing tables of unique sequence counts per sample, assigning taxonomy to sequences and and producing summary tables and plots, and computing standard alpha and beta diversity measures and summary plots to describe overall microbiome composition. These analysis are conducted using the core diversity analyses workflow of the QIIME 2 software package (https://qiime2.org) and with a standard workflow adapted from the “Moving pictures tutorials” (with some minor modifications)(https://docs.qiime2.org/2021.11/tutorials/moving-pictures/).
Custom bioinformatics support:
We provide custom bioinformatics support aligned with your research questions. We offer expertise in multi-omic analysis, combining data from assays such as 16S sequencing, multi-color flow cytometry and CyTOF panels, and metabolomics into an integrated whole. This integrated data can surface relationships across biological compartments, providing greater scientific insight than analysis of data one assay at a time. Our approaches include data visualization techniques and emphasize interpretability of results.
Some of these approaches are presented in PMID: 31429723, PMID: 33607938, PMID: 33811477, PMID: 31779604, PMID: 28764956, and PMID: 34006628. We are also able to investigate the application of recently published software to your data, subject to software availability.
Host: Microbe Experimental Validation
In order to support experimental validation of microbe:metabolite:immune phenotype relationships, we will 1) Provide services, consultation, and support in the expansion of anaerobic and aerobic bacteria and flow cytometry and density gradient based isolation of bacteria on a free/fee-for-service basis for RDRRC members. Provide free consultation and experimental design support in the assessment of mucosal immune cell function ex vivo or in response to bacteria using traditional flow cytometry and in vitro culture methods.
Anaerobic culture services:
The MIC provide services, consultation, and support in the expansion of anaerobic and aerobic bacteria. The MIC will aid with isolation and expansion of commensal and pathogenic bacteria at free and reduced fees. Core staff member Nusbacher has over 5 years’ experience in expanding both aerobic (e.g. commensal E. coli) and anaerobic (e.g. Bacteroides fluxus and Holdemanella biformis) bacteria. The Lozupone lab houses an anaerobic glove bag (Coy Type B 78” Vinyl), and a Coy Model 2000 incubator for growing bacterial cultures. We typically use a media called “Mega media” that supports the growth of most intestinal bacteria. This type of media is what is used to support “personalized culture collections” (i.e. collections of a diversity of bacteria all cultured from one particular fecal or mucosal sample). We also use various other specialized media to optimally grow certain bacteria or that are required for particular experimental designs (e.g. minimal growth media with various carbon sources added to test for growth requirements). The core will provide usage of the Coy chamber to individuals with prior experience/training in anaerobic culture and also provide training in anaerobic culture techniques or expansion of bacteria of interest on a fee for service basis.

Density Gradient-based isolation of bacteria:
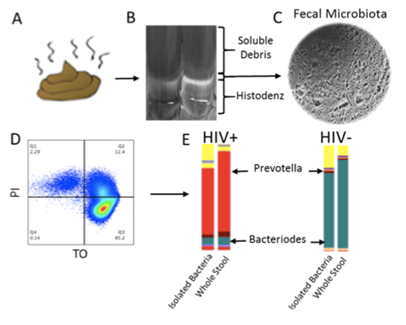
The Palmer lab has developed a method for isolating the full intact set of bacteria from fecal samples for use in in vitro assays. Specifically, a fecal slurry is made and layered on Histodenz and centrifuged. The bacterial layer is then collected and the viability and number of bacteria are analyzed by flow cytometry. These cell preparations can then be used to stimulate cell populations to investigate the immune-modulatory properties of fecal bacteria.
For an example see:
Neff CP, Krueger O, Xiong K, et al. Fecal Microbiota Composition Drives Immune Activation in HIV-infected Individuals. EBioMedicine 2018;30:192-202. doi: 10.1016/j.ebiom.2018.03.024 [published Online First: 2018/04/14]
The MIC will provide assistance with use of this technique for investigating host:microbe interactions that occur in rheumatologic disease.
Protocol for isolation of whole bacteria from fecal samples (FBCs). A fecal slurry is made and layered on Histodenz and centrifuged (B). The bacterial layer is collected and the viability and number of bacteria are analyzed by flow cytometry(D). Composition is analyzed by 16S rRNA sequencing and compared to whole stool (E).
Bacterial isolation with flow cytometry
A crucial goal of mucosal immunobiology research is identification of immunologically relevant bacteria. Using endogenous immunoglobulin coating of bacteria, usually by IgA but all isotypes can be present, bacteria targeted by the host immune system can be sorted from the uncoated fraction. Bacterial species of interest can also be flow sorted using antibodies or even polynucleotide probes, which leave the bacterial genetic material intact. Combining flow sorting with molecular tools like whole-genome amplification or PCR amplification of 16S rRNA allows for molecular taxonomic identification of specific bacterial populations. Using flow cytometry for isolation of bacteria is challenged by difficulties in isolating small cells which are difficult to distinguish from cellular debris. However, newer flow cytometry platforms, such as the Becton Dickenson FACSAria Fusion Cell Sorter with their upgraded fluidics system, reduce contamination of upstream and down-stream tubing after bacteria sorts and have high resolution, upgraded optics that allow for detection of smaller particles. Using the FACSAria Fusion the MIC will provide bacterial cell sorting from fecal matter, sputum, or any bacterial cells in suspension using antibodies or probes to RDRRC Investigators at a reduced cost . We have effectively employed these methods previously as described.
Palm NW, de Zoete MR, Cullen TW, et al. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell 2014;158(5):1000-10. doi: 10.1016/j.cell.2014.08.006 [published Online First: 2014/08/30]
The MIC will provide free consultation and a 25% discount off the standard fee for sorting at the Allergy and Clinical Immunology Flow Cytometry Facility run by Dr. Palmer. https://medschool.cuanschutz.edu/clinical-immunology/flow-core
Pricing
| Table 2: Fee Schedule for RDRRC Members and Affiliates | ||
|---|---|---|
| Service | Standard Fee | Member Fee |
| DNA extraction for 16S rRNA sequencing (per sample) | $56.37 | $47.91 |
| RNA extraction with Rnesasy PowerFecal Pro (per sample) | $67.01 | $31.46 |
| 16S rRNA amplicon sample prep (per sample) | $44.44 | $37.78 |
| 16S Amplicon Sequencing – 125+ samples | $13.90 | $11.82 |
| 16S Amplicon Sequencing – 90-124 samples | $21.88 | $18.60 |
| 16S Amplicon Sequencing – 15-30 samples | $35.49 | $30.17 |
| 16S Basic Analysis - per library | $1,650.00 | $1,402.50 |
| Metagenomics Basic Analysis (MetaG) - per library | $2,247.64 | $1,910.49 |
| Metatranscriptomics Basic Analysis (MetaT) - per library | $2,247.64 | $1,910.49 |
| Pre-sequencing consultation (per hour) | $75.00 | Free |
| Custom bioinformatics analysis (per hour) | $196.00 | $167.00 |
| Training in data analysis (per hour) | $196.00 | $167.00 |
| Use of anaerobic glove box (per month) | $75.00 | Free |
| Custom culturing support | $147.00 | $125.00 |
| Training in microbiologic culture (per hour) | $147.00 | $125.00 |
| Metabolomics Serum/Plasma | $193.00 | $158.26 |
| - Lipid fraction | $162.00 | $132.84 |
| - Aqueous fraction | $0.00 | $0.00 |
| Metabolomics Stool | $215.00 | $176.30 |
| - Lipid fraction | $184.00 | $150.88 |
| - Aqueous fraction | $100.00 | $82.00 |
| SCFA | $160.00 | $131.20 |
| Oxylipins | $125.00 | $102.50 |
| Tryptophan/Phenylalanine metabolism | $136.00 | $111.52 |
| Bile Acids | $136.00 | $111.52 |
| Endocannabinoids | $138.00 | $113.16 |
| Amino Acids | $98.00 | $80.36 |
| Nucleotides and Cofactors | $132.00 | $108.24 |
| NADome | $258.00 | $211.56 |
| TCA Cycle | $130.00 | $106.60 |
| Phytocannabinoids (THC, CBD, etc.) | $125.00 | $102.50 |
| Isolation of FBCs from feces | $50.00 | Free |
| Flow sorting of bacterial populations (per hour) | $125.00 | $97.00 |
| Cell isolation with microfluidics chip (per sample) | $50 | Free |
To learn more about any of the above services offered by the Mucosal Immunobiology Core (MIC), please complete this interest formform and a core representative will contact you to discuss your specific project needs.