Mucosal Immunobiology Core (MIC)
Overview
Compounding evidence exists for the role of microbiota and mucosal immunology in the pathogenesis of rheumatic diseases including rheumatoid arthritis (RA) and spondyloarthritis (SpA). The commensal microbiota modulates mucosal immune development and homeostasis through components of their cell envelope or capsule or through their metabolic activity.
These local processes can impact circulating and target organ immune phenotypes either through immune cell trafficking or translocation of microbial products. Thus, exploring mechanisms of host-microbe interactions pertinent to rheumatic disease involves in-depth phenotyping of microbial communities and host immune phenotypes in relevant tissues. Furthermore, although ‘omics technologies are valuable for producing mechanistic hypotheses regarding interactions between microbes and host, it is also important to validate these hypotheses in the lab.
One way to validate relationships predicted based on ‘omics analysis of human tissues is to stimulate specific human immune cell populations with cultured bacteria and/or their metabolites. This requires methods for isolating/cultivating bacteria and immune cell populations of interest. Accomplishment of these goals requires the integration of individuals with diverse expertise.
The Mucosal Immunobiology Core (MIC) aims to provide consultation, discounted services, and development of enhanced capabilities to support the prediction and validation of mechanisms of host:microbe interactions in biospecimens of importance to rheumatic disease pathogenesis, including the oral cavity, lower respiratory tract, intestinal mucosa, and cervicovaginal mucosa.
The MIC is committed to supporting both early career and established investigators in the University of Colorado (CU) Rheumatic Disease Research Resource Center (RDRRC), as well as assisting investigators new to the field of rheumatic disease to enhance overall research implementation and productivity.
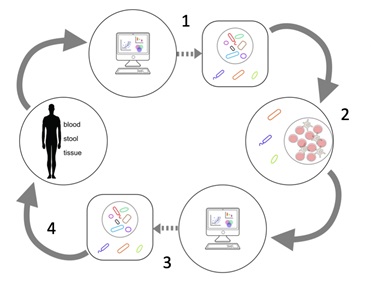 The MIC aims to facilitate an integrated approach where:
The MIC aims to facilitate an integrated approach where:
- Computational analysis of ‘omic data from human samples generates hypotheses regarding immune-modulatory bacteria of importance in rheumatic disease.
- These hypotheses are further validated and explored in in vitroimmune stimulation assays with human derived cells.
- Analyses of laboratory assays can lead to novel strategies of microbiome mediated interventions.
- These strategies can ultimately be tested in humans.
Our People
Catherine Lozupone, PhD: Core Director

Dr. Lozupone is an Associate Professor in the Division of Biomedical Informatics and Personalized Medicine in the Department of Medicine. Her research focuses on the complex community of microorganisms that inhabit the gastrointestinal tract. She has been heavily involved in the development of popular computational tools for microbial community analysis, such as the UniFrac algorithm for comparing microbial diversity among many samples using phylogenetic information. Dr. Lozupone currently runs an R01 funded research group that integrates integrative bioinformatics analysis of multi‘omic data with experimental confirmation using cultured anaerobic bacteria, immune stimulations, and gnotobiotic mice.
Kristine Kuhn, MD, PhD

Dr. Kuhn is a physician-scientist who is clinically trained as a rheumatologist and has extensive research training in mucosal immunity. Her independently funded research focuses on host:microbe interactions in the development of RA and SpA. She has published and ongoing studies mechanistically connecting microbiome, metabolome, and mucosal immune changes during the development of animal models of these diseases and in human cohorts. Additionally, Dr. Kuhn is the Director of the CU Gnotobiotic Mice Core Facility that she developed in 2015 to facilitate both her and the campus’ research in host:microbe interactions. Dr. Kuhn has extensive experience in integrating studies of the microbiome, metabolome, and immune cell function.
Kristen Demourelle, MD, PhD

Dr. Demoruelle is a physician-scientist who is clinically trained in rheumatology and has extensive research training in human subjects research and cellular phenotypes at mucosal sites that influence development of RA-related autoimmunity. Her independently funded laboratory focuses on understanding the earliest steps of RA development, specifically the development of RA-related autoantibodies prior to the onset of joint disease during the ‘pre-clinical’ phase of RA, and sex differences in this process. She has published and ongoing studies that identify novel biomarkers and cellular phenotypes in the lung and female genital tract associated with RA-related autoimmunity and RA-related lung disease. Her lab also investigates mechanistic pathways by which neutrophils and mucosal bacteria can contribute to autoimmunity and disease progression, with a specific focus on microbial-induced and non-microbial-induced neutrophil extracellular trap (NET) formation and clearance.
Brent Palmer, PhD

Dr. Palmer is an Associate Professor in the Department of Medicine, Division of Allergy and Clinical Immunology. Dr. Palmer’s laboratory studies the interactions between commensal microbiota and the mucosal immune system (gut and lung) and the impact of these interactions of HIV disease progression.Dr. Palmer is also the Director of the Allergy and Clinical Immunology research and clinical CAP/CLIA accredited Flow Cytometry facility and has 30 years of flow cytometry experience with > 85 publications in the field of clinical immunology and host-microbe interactions. In addition, Dr. Palmer has extensive experience in generation and analysis of complex multi-color flow cytometry and CyTOF data and experimental validation of microbe:metabolite:immune phenotype relationships via in vitro cell stimulations using individual bacterial species or whole bacterial communities.
Nichole Reisdorph, PhD

Dr. Reisdorph is a Professor in the Department of Pharmaceutical Sciences. Dr. Reisdorph has been a Mass Spectrometry Facility Director for over 15 years, has NIH funding in several areas of metabolomics/small molecule analysis, and directs hands on workshops in metabolomics and proteomics. Dr. Reisdorph also has extensive experience in understanding the role of bioactive lipids in inflammatory disease such as asthma. Dr. Reisdorph pioneered the use of sample type specific databases in improving confidence in compound identification.
Maggie Stanislawski, PhD

Dr. Stanislawski is an Assistant Professor in the Division of Biomedical Informatics and Personalized Medicine in the Department of Medicine. She is a molecular epidemiologist, and her work aims to understand the role of the gut microbiome and related molecular profiles in health and disease, specifically obesity, cardiometabolic disease and inflammatory conditions. In addition to her PhD in epidemiology, she has an MSc in statistics, and she has many years of experience as a Biostatician, with expertise in study design for diverse biomedical research questions and statistical methods for the analysis of microbiome and other ‘omics data.
Nichole Nusbacher MS

Ms. Nusbacher is a MS graduate with significant experience in microbiology and molecular biology. Since 2015, she has been actively working on microbiome-related projects for Dr. Lozupone. She has substantial experience in the next generation sequencing of microbiome samples on the MiSeq platform, microbiome data analysis, and anaerobic culture.
Michael Armstrong, BS

Michael Armstrong directs the quantitative small molecule analysis at the Mass Spectrometry Core and has over 20 years of mass spectrometry and analytical chemistry experience.Previous to working in the School of Pharmacy Mass Spectrometry Facility, Michael worked at the National Jewish Health Mass Spectrometry Core facility for 9 years. His professional interests include mass spectrometry based metabolomics and analytical chemistry. Michael has developed over 15 targeted mass spectrometry assays that cover over 250 small molecules.
Richard Reisdorph, PhD

Rick received his PhD in Molecular Biology and Biochemistry from the University of South Dakota School of Medicine and was a fellow with Peter Vogt at the Scripps Research Institute in La Jolla, CA. His work includes developing quantitative metabolomics and proteomics techniques and bioinformatics. He is currently the Associate Director of the SSPPS Mass Spectrometry Core Facility and Directs the Metabolomics efforts.
Janet Siebert, PhD

Dr. Siebert offers over 25 years of experience integrating and analyzing complex data. She has co-authored peer reviewed publications based on data generated by flow cytometry and CyTOF, Luminex, MSD, 16S rRNA sequencing, metabolomics, DNA methylation arrays, and protein arrays. The thesis for her MS in Computer Science was titled “Knowledge Discovery in Flow Cytometry Data.” More recently, she has developed informatics techniques to study microbiome-immune system interplay, focusing on interactive data visualization and supporting cross-disciplinary communication.