Beat Vögeli, PhD
Associate Professor

Contact Information:
Fax: (303) 724-1627
E-mail: Beat.Vogeli@cuanschutz.edu
Graduate Program Memberships:
NMR spectroscopy for the elucidation of conformation and communication networks within and between proteins, nucleic acids and small compounds
Recent breakthroughs in nuclear magnetic resonance (NMR) spectroscopy, X-ray crystallography, cryo-electron microscopy and other biophysical techniques have reshaped our views dynamics and function of proteins and nucleic acids. We are beginning to grasp a more holistic and fascinating reality in which many, if not the majority of the proteins and nucleic acids are operative as communication hubs that are interconnected with many others. Domain architecture is defined by the 3D average structure, but biomolecules are inherently dynamic systems. Our dynamic picture is still largely incomplete because no adequate methods are available to detect concerted conformational transitions at atomic resolution. These transitions, however, form the structural basis for relaying information from one molecular site to another (so-called allostery).
NMR spectroscopy is one of the principal techniques to answer fundamental questions addressing protein/nucleic acid function due to its analytical potential at an atomic level. Using NMR, we contribute to the understanding of communication networks by
modeling realistic ensembles of structures, identification of allosteric mechanisms, studying folding and interactions with other proteins, nucleic acids or small ligands. An important pillar of this program is the establishment of our recently developed
exact nuclear Overhauser enhancement (eNOE) methodology for a realistic representation of molecular spatial sampling as a standard tool in the NMR/structural biology community.
Exact NOE methodology
Standard structure determination by NMR spectroscopy makes use of the abundant number of experimentally readily accessible NOE rates. These rates are typically employed in a semi-quantitative manner because the measurement is obscured by spin diffusion, low signal-to-noise ratio and technical limitations. We have proposed a novel method for the exact measurement of NOEs (eNOEs). Distances obtained from eNOEs can have an experimental error of only 0.1 Å. The collection of thousands of eNOEs throughout a biomacromolecule serves as an excellent probe towards a more complete representation of the structural landscape.
We have developed a program that guides the analysis of eNOEs and extraction of distances for structure calculation (eNORA, available either in Matlab format or integrated into CYANA). We have applied the method to various proteins and are also extending the method to RNA and DNA (see eNOE structure gallery).
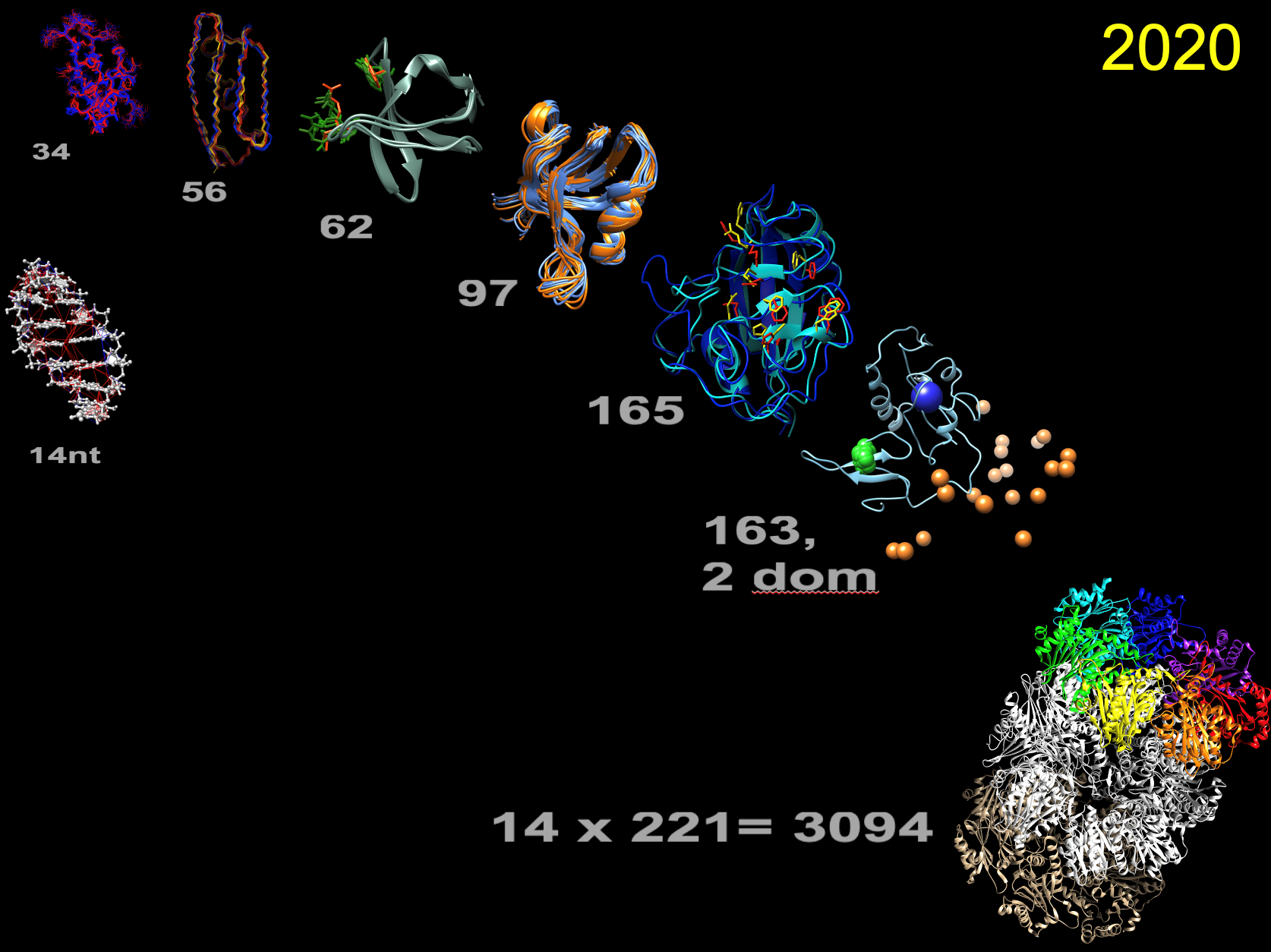
Figure 1 eNOE structure gallery. Proteins sorted by increasing residue lengths (grey numbers): WW domain of human Pin1, 3rd immunoglobulin binding domain of protein G (GB3), sarcoma homolog 3 (SH3) of chicken alpha-spectrin, PDZ2 of human tyrosine phosphatase 1E (hPTP1E), human cyclophilin A, two-domain full-length human Pin1, 20S core particle proteasome of T. acidophilium (only distances). 14-nucleotide UUCG RNA tetraloop (left).
Systems studied
Motor adapters in complex dynamic protein assemblies associated with the motor cytoplasmic dynein
Cytoplasmic dynein 1 (‘dynein’), a multi-protein complex of 1.2 megadaltons, is the predominant microtubule minus-end-directed motor in animals and humans. Dynein participates in a wide range of cellular activities, ranging from the transport of proteins, RNA, and vesicles to nuclear migration and cell division. Specific adapter proteins link dynein to cargo and activate the motor by forming a ternary complex with dynein and its essential co-activator dynactin. Mutations in dynein, as well as in its co-factors and adapters, have been linked to multiple neurodegenerative diseases. Several adapters have been shown to interact with the C-terminal tail of dynein light intermediate chain (LIC), which is predicted to be disordered, and mutations in the interacting segments have been implicated in spinal muscular atrophy.
It is our aim to use NMR-based experiments with the LIC C-terminal tail to provide molecular insight into how the elongated, flexible scaffold engages with functionally diverse dynein adapters. This project is realized in collaboration with Reto Gassmann at the Institute for Molecular and Cell Biology (IBMC) in Porto.
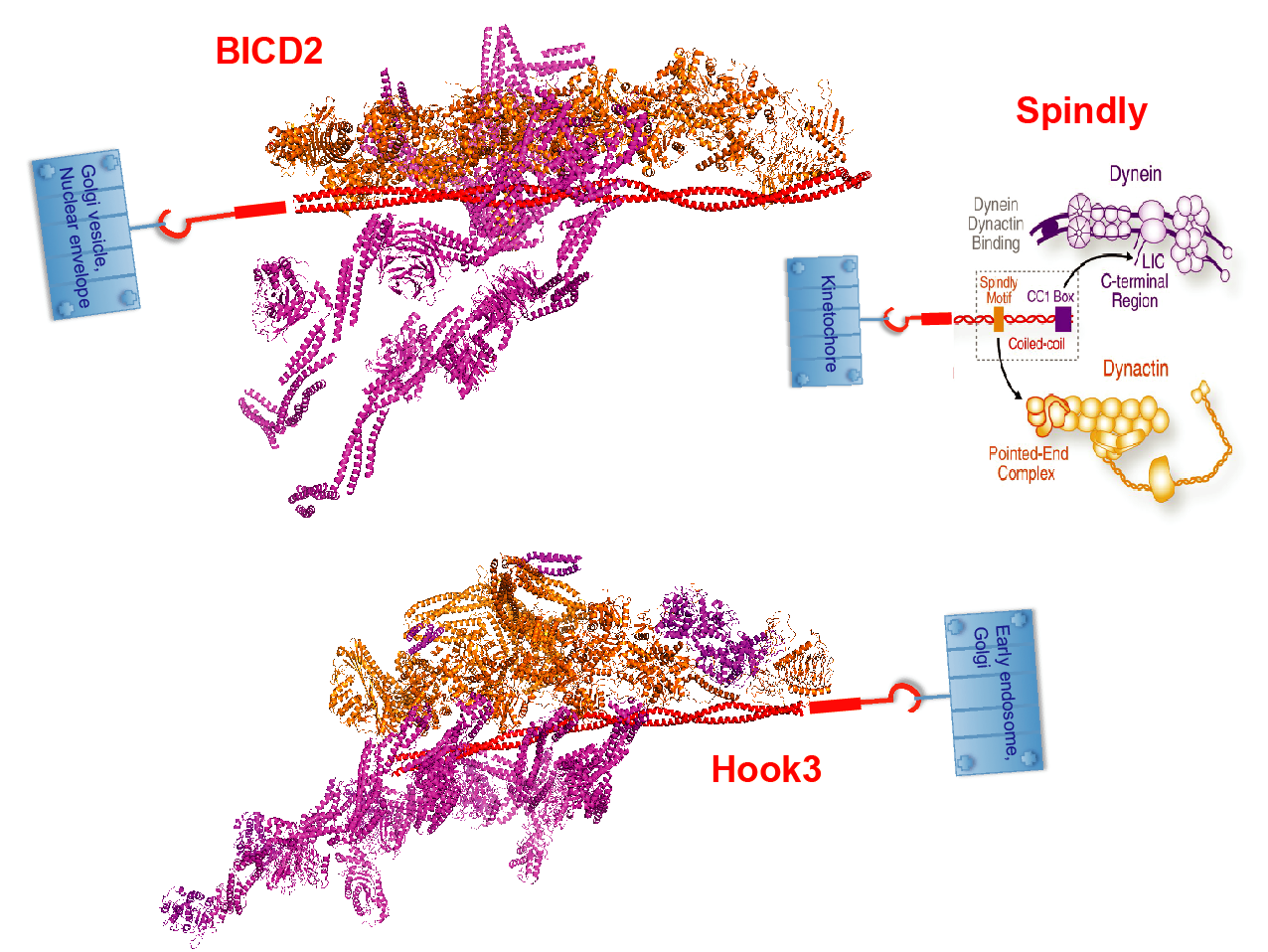
Figure 2 Dynein-associated transport machineries have various adapters and cargoes. Dynein/dynactin complexes (purple/orange) are bound to the three example adapter proteins BicD2, Spindly and Hook (red). The N-termini of the adapters are drawn as ribbons, and the cargo-binding C-termini are indicated by hooks attached to their respective cargoes.
Structural landscape of Pin1 allostery and WW domain folding
The human peptidyl-prolyl cis/trans isomerase (PPIase) Pin1 exhibits an intricate example of allostery in a two-domain protein with high biological significance. It is involved in the regulation of mitosis, protection against Alzheimer’s disease, increase of hepatitis C infection and it is overexpressed in many human cancer cells. The catalytic domain isomerizes pS/T-P containing peptide motifs in a large variety of ligands. Not only the structure but also the dynamics of the native 34-residue WW-domain seems to be responsible for the specificity of binding. Interestingly, the WW domain transmits information from its binding site to the spatially separated catalytic PPI site. The underlying mechanism remains a riddle. In our initial studies of the apoform WW domain, we identified a dynamic network spanning most of the minicore and loop 2.
It is our aim to get an atomic-resolution view of the allosteric functionality of the entire Pin1, and the folding pathway(s) of its WW domain. We use and develop further eNOE, standard NMR and electron paramagnetic resonance (EPR, in collaboration with Gunnar Jeschke, ETH Zürich) spectroscopy.
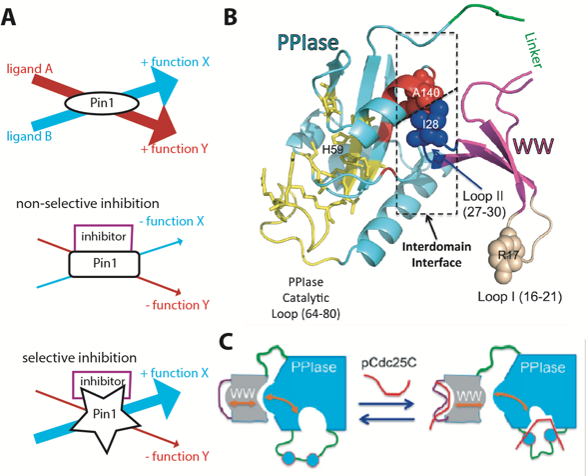
Figure 3 Pin1 is a two-domain pathway hub. A) Selective allostery of Pin1. B) Both domains bind pS/T-P motifs: the WW domain (purple/brown/blue) and the PPIase domain (cyan/yellow/red), which catalyzes cis-trans isomerization. Shown is the closed state, where the red and blue atoms form an interdomain interface. Adapted from Peng 2015, Biophys Rev 7, 239. C) Pin1 is in an equilibrium between closed and open conformations, which is shifted upon ligand binding via an allosteric circuit (orange arrows). Adapted from Wang et al 2015, Structure 23, 2224.
Functional dynamics in RNA
Most biological processes involve non-coding RNA (ncRNA), such that RNA is now established as one of the key players in the regulation of gene expression. RNA generally alters gene expression by shifting between distinct three-dimensional architectures
that differentially interact with other molecules. Using NMR and further biophysical methods, we aim to visualize the dynamic aspects of such conformational changes. We are interested in loop dynamics, ligand-induced conformational changes, and the
dynamics of Z-RNA formation (in-house collaboration with Quentin Vicens and Jeffrey Kieft).
Allostery in protein-RNA interaction
Non-coding RNA (ncRNA) has recently generated much interest due to its ability to sequester protein. In the bacterial Csr/Rsm system, which is the most general global post-transcriptional regulatory system responsible for bacterial virulence, mRNA such as CsrB or RsmZ initiates translation through sequestration of homodimeric CsrA-type proteins bound to ribosome-binding sites of messenger RNAs. Our collaborator Frédéric Allain (ETH Zürich) has shown that in the case of Pseudomonas fluorescens, RsmE protein dimers assemble sequentially and specifically onto ribonucleoprotein structures, when binding the non-coding RNA RsmZ. The binding cascade of RsmE dimers to one RsmZ is cooperative but binding of the second stem-loop (SL) happens via a negative allosteric effect. Our preliminary NMR analysis of the binding of two isolated SL2 to RsmE suggests a mechanism of inter-subunit allostery in RsmE that involves two distinct intermediate states in slow exchange.
It is our aim to reveal the allosteric mechanism underlying SL2 binding to RsmE at atomic level using eNOE-based and established NMR techniques. On a more fundamental level, the elucidation of this allosteric mechanism may uncover more general principles
on how RNA binding domains in tandem or homodimeric RNA binding proteins fine tune their RNA binding affinity.
Ligand-protein interaction studies for drug design
NMR is one of the leading techniques in the field of drug discovery due to its high sensitivity. We are collaborating with various groups in Anschutz Medical campus to study ligand-protein interaction as a step towards the design of therapeutic agents targeting different diseases.

| Pictures | First Name | Last Name | Job Title | |
|---|---|---|---|---|
 | Morkos | Henen | Assistant Research Professor | morkos.henen@cuanschutz.edu |
 | Jeff | Krall | Graduate Student | jeffrey.krall@cuanschutz.edu |
 | Parker | Nichols | Graduate Student | |
 | Natasia | Paukovich | Graduate Student |
Pulse Sequence Code
Bruker pulse sequences for the measurement of dipolar cross-correlated relaxation between consecutive HN-N
Reference: B. Vögeli, Cross-correlated relaxation rates between protein backbone H-X dipolar interactions, 2017, J Biomol NMR, 67, 211-232
Bruker pulse sequences for the measurement of dipolar cross-correlated relaxation between consecutive HA-CA
Reference: B. Vögeli, Cross-correlated relaxation rates between protein backbone H-X dipolar interactions, 2017, J Biomol NMR, 67, 211-232
Bruker pulse sequences for the measurement of dipolar cross-correlated relaxation between intraresidual and sequential HN-N and HA-CA by MMQ
Reference: B. Vögeli, Cross-correlated relaxation rates between protein backbone H-X dipolar interactions, 2017, J Biomol NMR, 67, 211-232
Bruker pulse sequences for the measurement of dipolar cross-correlated relaxation between HN-N and HB-CB in Thr, Ile, Val
Reference: R.B. Fenwick & B. Vögeli, Detection of correlated protein backbone and side-chain angle fluctuations, 2017, ChemBioChem, 18, 2016-2021, DOI: 10.1002/cbic.201700312
Bruker pulse sequence for the measurement of eNOE buildups between methyl groups
Reference: C.N. Chi, D. Strotz, R. Riek & B. Vögeli, NOE-derived methyl distances from a 360 kDa proteasome complex, 2018, Chem Eur J, 24, 2270-2276, DOI: 10.1002/chem.201705551
Varian pulse sequence for the measurement of HA-HB(2,3) J-couplings
Reference: A. Born, M.A. Henen, P. Nichols, J. Wang, D.N. Jones, B. Vögeli, Efficient stereospecific Hβ2/3 NMR assignment strategy for mid-size proteins, 2018, Magnetochemistry, 4, 25, doi.org/10.3390/magnetochemistry4020025
Bruker pulse sequence for the measurement of eNOE buildups between all protons in a 15N/13C isotope-labeled protein via 3D [1H(F3),1H]-NOESY-[simultaneous 15N/13C(F2),1H(F1)]-HSQC
Reference: B. Vögeli, P. Güntert & R. Riek, Multiple-state ensemble structure determination from eNOE spectroscopy, 2013, Mol Phys, 111, 437-454, DOI: 10.1080/00268976.2012.728257
More Bruker pulse sequences can be found at:
http://n.ethz.ch/~bvoegeli/beat.html
Data Analysis Programs
eNORA2 MatLab code for the extraction of exact cross-relaxation rates and upper- and lower distance limits from NOESY buildups
Reference: D. Strotz, J. Orts, C. Chi, R. Riek, B. Vögeli, eNORA2 exact NOE analysis program, 2017, J Chem Theory Comput, DOI: 10.1021/acs.jctc.7b00436.
The first comprehensive book on the fundamentals, analysis and applications of RDCs is out now!
Residual Dipolar Couplings: Principles and Applications

Royal Society of Chemistry book series New Developments in NMR, 402 pages
Residual dipolar couplings (RDCs) serve as prominent NMR tools for eliciting structural and dynamic details in both small compounds and complex macromolecules. Edited by Lishan Yao and Beat Vögeli, this book offers a comprehensive overview of RDCs, from fundamental principles to advanced applications in organic molecules and biomolecules. Renowned experts in the field delve into the latest advancements in RDC measurement and analysis.
Whether readers are seasoned professionals or aspiring students, this book equips them with a robust comprehension of RDC fundamentals and their potential integration into their research endeavors.