Mamuka Kvaratskhelia, PhD

Professor of Medicine
Division of Infectious Diseases
University of Colorado School of Medicine
12700 E. 19th Avenue, Box B-168
Research Complex 2, Room 11002
Aurora, CO 80045
Phone: 303-724-3862
Email: MAMUKA.KVARATSKHELIA@CUANSCHUTZ.EDU
The research in our laboratory focuses on new generation HIV-1 inhibitors and virus-host interactions. We employ complementary biochemistry, structural biology, pharmacology, molecular biology and virology approaches.
Long-acting HIV-1 capsid inhibitors. Antiretroviral therapies (ART) have transformed the once deadly HIV/AIDS disease into a manageable, chronic infection. Yet, there are a number of pressing problems associated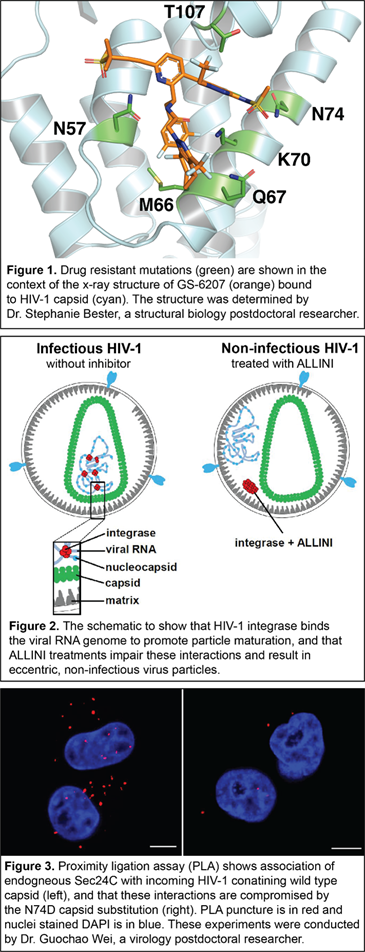 with current ARTs, including the necessity of daily administration of HIV-1 medications, suboptimal treatment adherence, and the emergence of drug-resistant viral phenotypes. This created a need for a forthcoming new class of long-acting antiretroviral agents that target clinically unexploited viral proteins and could transform the care of people living with HIV-1 by mitigating the above problems. HIV-1 capsid protein, which plays multiple essential roles during the virus life cycle, is an unprecedented clinical target. GS-6207 (Lenacapavir, Gilead Sciences) is a recently discovered, first-in-class, long-acting, and ultra-potent HIV-1 capsid inhibitor. This experimental drug, which is undergoing Phase II/III clinical trials, could offer up to a six-month dosing interval. Our recent studies have elucidated structural insights into the multimodal, potent antiviral activity of GS-6207 (1). Furthermore, our high-resolution x-ray structure of GS-6207 bound to a HIV-1 capsid hexamer enabled us to map drug-resistant mutations, which were observed in cell culture and patients, in close proximity to the GS-6207 binding site (Figure 1). We are currently investigating the underlying mechanisms for these drug resistance mutations. This information will be critical for the rational design of second-generation HIV-1 capsid inhibitors with a higher genetic barrier to resistance.
with current ARTs, including the necessity of daily administration of HIV-1 medications, suboptimal treatment adherence, and the emergence of drug-resistant viral phenotypes. This created a need for a forthcoming new class of long-acting antiretroviral agents that target clinically unexploited viral proteins and could transform the care of people living with HIV-1 by mitigating the above problems. HIV-1 capsid protein, which plays multiple essential roles during the virus life cycle, is an unprecedented clinical target. GS-6207 (Lenacapavir, Gilead Sciences) is a recently discovered, first-in-class, long-acting, and ultra-potent HIV-1 capsid inhibitor. This experimental drug, which is undergoing Phase II/III clinical trials, could offer up to a six-month dosing interval. Our recent studies have elucidated structural insights into the multimodal, potent antiviral activity of GS-6207 (1). Furthermore, our high-resolution x-ray structure of GS-6207 bound to a HIV-1 capsid hexamer enabled us to map drug-resistant mutations, which were observed in cell culture and patients, in close proximity to the GS-6207 binding site (Figure 1). We are currently investigating the underlying mechanisms for these drug resistance mutations. This information will be critical for the rational design of second-generation HIV-1 capsid inhibitors with a higher genetic barrier to resistance.
Allosteric HIV-1 integrase inhibitors. We are investigating allosteric HIV-1 integrase inhibitors (ALLINIs) for their future clinical development (2,3). Our structure-activity relationship studies uncovered the principal antiviral mode of action of these inhibitors and delineated the importance of HIV-1 integrase multimerization as a novel, attractive therapeutic target (4,5). We are currently developing highly potent pyridine-based inhibitors by rationally modifying their quinoline-based predecessors (3,5). Our lead pyridine-based inhibitor KF116 exhibits substantially enhanced, sub-nM antiviral activity against a clinically relevant dolutegravir resistant mutant virus indicating exciting potential benefits for combining these two classes of inhibitors for treating HIV-1 infected patients (3).
Furthermore, we exploit ALLINIs as powerful investigational tools for better understanding of HIV-1 molecular biology. We have found that ALLINIs induce aberrant integrase multimerization in viral particles during maturation, which consequently impairs integrase binding to the viral RNA genome and results in eccentric, non-infectious virions with ribonucleoprotein complexes being displaced outside of the protective capsid core (Figure 2). In turn, these findings allowed us to discover an unexpected, non-catalytic role of integrase in HIV-1 biology, which includes its ability to bind and localize the viral RNA genome within the protective cone-shaped capsid core during virion maturation (2). Our current efforts are focused on dissecting structural and mechanistic determinants for highly specific interactions of HIV-1 integrase with select sites on the viral RNA genome.
Virus-host interactions. As an RNA virus with a limited genome size, HIV-1 has evolved to exploit host factors to traffic across the cytoplasm, through the nuclear pore and into the nucleus to ensure productive infection. A virus capsid shell provides the main interface with the complex cellular milieu post-entry. However, the identity and roles of capsid-binding cellular factors are poorly deciphered at present. Moreover, it is not clear how viral capsid binds selectively to diverse and seemingly unrelated host proteins. We have recently discovered Sec24C as an HIV-1 host dependency factor crucial for virus spreading infection (6). Sec24C directly and specifically interacts with hexameric capsid lattices, and enhances stability of HIV-1 capsid in the cytoplasm during virus ingress (Figure 3). Furthermore, our studies have uncovered a network of novel cellular capsid-binding containing proteins residing in different cellular compartments. We are currently investigating underlying mechanisms for avid and selective recognition of incoming HIV-1 capsid by these cellular co-factors.
Another project in our lab focuses on interactions of retroviral integrases with cognate cellular cofactors. Specifically, we want to better understand how LEDGF/p75, a principal cellular binding partner of lentiviral integrases, navigates HIV-1 preintegration complexes to active genes during integration (7-9). Moreover, our earlier studies have identified bromodomain and extraterminal domain (BET) proteins (Brd2, 3, 4) as principal cellular-binding partners of gammaretroviral murine leukemia virus (MLV) integrase and demonstrated their importance for targeting MLV integration to transcription start sites (10). These findings inform ongoing efforts to develop safer retroviral vectors for human gene therapy.
1. Bester, S. M., Wei, G., Zhao, H.,
Adu-Ampratwum, D., Iqbal, N., Courouble, V. V., Francis, A. C., Annamalai, A.
S., Singh, P. K., Shkriabai, N., Van Blerkom, P., Morrison, J., Poeschla, E.
M., Engelman, A. N., Melikyan, G. B., Griffin, P. R., Fuchs, J. R., Asturias,
F. J., and Kvaratskhelia, M. (2020) Structural and mechanistic bases for a
potent HIV-1 capsid inhibitor. Science
370, 360-364
2. Kessl,
J. J., Kutluay, S. B., Townsend, D., Rebensburg, S., Slaughter, A., Larue, R.
C., Shkriabai, N., Bakouche, N., Fuchs, J. R., Bieniasz, P. D., and
Kvaratskhelia, M. (2016) HIV-1 Integrase Binds the Viral RNA Genome and Is
Essential during Virion Morphogenesis. Cell
166, 1257-1268 e1212
3. Koneru,
P. C., Francis, A. C., Deng, N., Rebensburg, S. V., Hoyte, A. C., Lindenberger,
J., Adu-Ampratwum, D., Larue, R. C., Wempe, M. F., Engelman, A. N., Lyumkis,
D., Fuchs, J. R., Levy, R. M., Melikyan, G. B., and Kvaratskhelia, M. (2019)
HIV-1 integrase tetramers are the antiviral target of pyridine-based allosteric
integrase inhibitors. eLife 8
4. Jurado,
K. A., Wang, H., Slaughter, A., Feng, L., Kessl, J. J., Koh, Y., Wang, W., Ballandras-Colas,
A., Patel, P. A., Fuchs, J. R., Kvaratskhelia, M., and Engelman, A. (2013)
Allosteric integrase inhibitor potency is determined through the inhibition of
HIV-1 particle maturation. Proc Natl Acad
Sci U S A 110, 8690-8695
5. Sharma,
A., Slaughter, A., Jena, N., Feng, L., Kessl, J. J., Fadel, H. J., Malani, N.,
Male, F., Wu, L., Poeschla, E., Bushman, F. D., Fuchs, J. R., and
Kvaratskhelia, M. (2014) A new class of multimerization selective inhibitors of
HIV-1 integrase. PLoS Pathog 10,
e1004171
6. Rebensburg,
S. V., Wei, G., Larue, R. C., Lindenberger, J., Francis, A. C., Annamalai, A.
S., Morrison, J., Shkriabai, N., Huang, S. W., KewalRamani, V., Poeschla, E.
M., Melikyan, G. B., and Kvaratskhelia, M. (2021) Sec24C is an HIV-1 host
dependency factor crucial for virus replication. Nat Microbiol
7. Eidahl,
J. O., Crowe, B. L., North, J. A., McKee, C. J., Shkriabai, N., Feng, L.,
Plumb, M., Graham, R. L., Gorelick, R. J., Hess, S., Poirier, M. G., Foster, M.
P., and Kvaratskhelia, M. (2013) Structural basis for high-affinity binding of
LEDGF PWWP to mononucleosomes. Nucleic
Acids Res 41, 3924-3936
8. Singh,
P. K., Plumb, M. R., Ferris, A. L., Iben, J. R., Wu, X., Fadel, H. J., Luke, B.
T., Esnault, C., Poeschla, E. M., Hughes, S. H., Kvaratskhelia, M., and Levin,
H. L. (2015) LEDGF/p75 interacts with mRNA splicing factors and targets HIV-1
integration to highly spliced genes. Genes
Dev 29, 2287-2297
9. Passos,
D. O., Li, M., Yang, R., Rebensburg, S. V., Ghirlando, R., Jeon, Y., Shkriabai,
N., Kvaratskhelia, M., Craigie, R., and Lyumkis, D. (2017) Cryo-EM structures
and atomic model of the HIV-1 strand transfer complex intasome. Science 355, 89-92
10. Sharma, A., Larue, R. C., Plumb, M. R.,
Malani, N., Male, F., Slaughter, A., Kessl, J. J., Shkriabai, N., Coward, E.,
Aiyer, S. S., Green, P. L., Wu, L., Roth, M. J., Bushman, F. D., and
Kvaratskhelia, M. (2013) BET proteins promote efficient murine leukemia virus
integration at transcription start sites. Proc
Natl Acad Sci U S A 110, 12036-12041