Shared Content Block:
Styles for CDB Faculty Pages
Tom Evans, Ph.D.
Associate Professor

| tom.evans@cuanschutz.edu | |
| 303-724-3414 | |
| Ph.D. — University of Wisconsin, 1988 |
Graduate Program Affiliations:
Control of stem cell development by RNA/protein complexes
.jpg?sfvrsn=22d5bdba_2)
Figure 1. In the C. elegans adult gonad, germline stem cells generate oocytes by a carefully orchestrated process. Oocytes then become embryos where polarized (asymmetric) cell division generates various precursors and stem cells
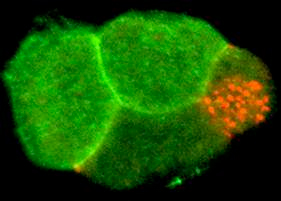
Figure 2. A 4-cell embryo stained for the membrane receptor Notch (green) and P granules (red). Notch is localized to anterior cells where it controls anterior cell fates. P granules are localized to a single posterior cell, the germline stem cell, at each cell division. Other cell fate regulators have distinct expression patterns. The behavior of mRNP complexes is dynamic; their composition, structure, localization change during development. These complexes regulate germ cell differentiation and stem cell development in the embryo. Future research in the lab will aim to understand how mRNP complexes are regulated in living cells during development, to explore how they control patterns of protein translation, and to identify novel proteins and RNAs that mediate this control.
Animal development depends on the precise timing and location of protein activities. Regulation of gene transcription has long been considered central to this process. However, recent work argues that many critical mRNAs are also tightly controlled in the cytoplasm after they leave the nucleus, leading to specific patterns of protein translation and localization. Cytoplasmic mRNA regulation is very important during oogenesis, early embryogenesis, and during stem cell divisions that occur throughout development. The mechanisms that regulate cytoplasmic mRNAs and that tie this control to the development of living cells are poorly understood. Recent studies hint that the mechanisms are similar in many animals and that they are important for various human diseases.
We are exploring this problem in the small nematode worm, Caenorhabditis elegans. In the C. elegans adult gonad, germline stem cells generate oocytes by a carefully orchestrated process. Oocytes then become embryos where polarized (asymmetric) cell division generates various precursors and stem cells (Figure 1).
These events lead to precise localization of developmental regulators in the embryo (Figure 2). mRNAs for these regulators are produced by germ cells in the mother and are packaged into oocytes and embryos. We have found that several different RNA-binding proteins function at specific times and places to control numerous mRNAs during development.
Interestingly, each of these proteins has human relatives, but their functions in people are not known. These proteins and their regulated mRNAs form various large mRNA/protein complexes (mRNPs) in the cytoplasm of oocytes and embryos (the red particles in Figure 2 are one type of mRNP complex).
Why do this research in C. elegans? The genes that control C. elegans biology are similar to human genes, several of which have been tied to cancer, autoimmune disease, nervous system disorders, and other diseases. Sophisticated techniques can be combined in C. elegans to yield new discoveries. Detailed features of the worm genome have been determined. Gene function can be knocked out by mutation or by the simple technique of RNA interference. The function and expression of these genes can be analyzed in living animals using high-resolution microscopy. mRNP complexes can be isolated and their components identified using mass spectrometry and microarray technology. Recombinant genes, mRNAs, and proteins can be re-introduced into these animals. The C. elegans system allows the discovery of new genes important in human biology and disease, and detailed studies of their function and regulation.
View Dr. Evans' Latest Publications in PubMed
Noble, S.L., Allen, B., Goh, L.K., Nordick, K., Evans, T.C. (2008) Maternal mRNAs are regulated by diverse P body-related mRNP granules during early Caenorhabditis elegans development. J. Cell Biology 182, 559-572.
Lublin, A.L. and Evans, T.C. (2007) The RNA-binding proteins PUF-5 and PUF-6/7 reveal multiple systems for maternal mRNA regulation during C. elegans oogenesis Developmental Biology 303, 635-649.
Barbee, S.A., and Evans, T.C. (2006) The Sm proteins regulate germ cell specification during early C. elegans embryogenesis. Developmental Biology. 291, 132-143.
Evans, T.C. and Hunter, C.P. (2005). Translational control of maternal RNAs. Wormbook, ed. The C. elegans Research Comm. Wormbook, doi/10.1895/wormbook.1.34.1, http://www.wormbook.org/
Evans, T.C. (2006) Transformation and microinjection. WormBook, ed. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.108.1, http://www.wormbook.org.
Marin, V.M. and Evans, T.C. (2003). Translational repression of a C. elegans Notch mRNA by the STAR/KH domain protein GLD-1. Development. 130, 2623-2632.
Barbee, S.L., Lublin, A., and Evans T.C. (2002). A Novel function for the Sm proteins in germ granule localization during C. elegans embryogenesis. Current Biology. 12, 1502-1506.
Goodwin, E.B., and Evans, T.C. (1997). Translational control of development in C. elegans. Seminars Cell Dev Biol. 8: 551-559.
Evans, T.C., S. Crittenden, V. Kodoyianni, and J. Kimble. (1994). Translational control of maternal glp-1 mRNA establishes an asymmetry in the C. elegans embryo. Cell 77, 183-194.
Crittenden, S., E. Troemel, T.C. Evans and J. Kimble. (1994). GLP-1 is localized to the mitotic region of the C. elegans germline. Development 120, 2901-2911.